Primitive Synteny of Vertebrate Major Histocompatibility Complex Class I and Class II Genes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Korea, Republic Of
Ministry for Food, Agriculture, Forestry and Fisheries National Plan of Action for the Conservation and Management of Sharks The Republic of Korea August, 2011 MIFAFF Ministry for Food, Agriculture, Forestry and Fisheries I. Introduction ············································································ 2 II. Current State of Shark Management in Korea ························· 3 2.1. Spatial Distribution of Sharks and Related Fisheries ····················· 3 2.2. Shark Catch Statistics ··································································· 7 2.3. Domestic and International Trade of Sharks ································· 8 2.4. Management of Sharks ································································· 9 III. National Plan of Action for the Conservation and Management of Sharks · 10 3.1. Objectives and Scope of National Plan of Action for Sharks······· 10 3.2. Data Collection, Analysis and Assessment ··································· 11 3.3. Conservation and Management Measures ···································· 13 3.4. Monitoring, Control and Surveillance ··········································· 15 3.5. Maximizing the Utilization of Sharks Caught ······························· 16 3.6. Research and Development ························································· 17 3.7. Public Information and Promotion ··············································· 18 3.8. International Cooperation ····························································· 19 3.9. Other Issues ················································································ -

Download the Full Article As Pdf ⬇︎
LocalLockdown Diving — Dives Found in Contributors' Backyards Text and photos by Andrey Bizyukin, Larry Cohen, Brent Durand, Dmitry Efremychev, Jennifer Idol, Kate Jonker, Matthew Meier, Pete Mesley, Don Silcock, Olga Torrey and Martin Voeller As many divers face travel restrictions during the coronavi- rus pandemic, our contributors highlight the often overlooked or unsung yet intriguing div- ing that can be found in one's own backyard. X-Ray Mag contributors share their favorite local haunts—from a spring-fed Texan lake to a quarry and a sinkhole in Russia to the tem- perate waters off New Zealand, Japan, South Africa, New Jersey and Northern California to the subtropical waters of Southern California and Sydney, Australia—where they captured compelling underwater images. 58 X-RAY MAG : 101 : 2020 EDITORIAL FEATURES TRAVEL NEWS WRECKS EQUIPMENT BOOKS SCIENCE & ECOLOGY TECH EDUCATION PROFILES PHOTO & VIDEO PORTFOLIO MATTHEW MEIER feature Local Dives School of opaleye and garibaldi among sea grass and feather boa kelp. PREVIOUS PAGE: School of juvenile senorita fish in the kelp and sea grass beds Bat Ray Cove, San Clemente Island, Soupfin (tope) sharks can be seen swimming in California, USA the shallows and among the giant kelp, along with schools of blacksmith and jack mackerels. Text and photos by Matthew Meier California sea lions will swoop through intermittently, and the occasional harbor seal Thankfully, local diving is still possible during will play peak-a-boo in the kelp. the pandemic, and while this dive site requires Under the boat is a sandy bottom where boat access, it is still one of my favorites. -

Investigation Report of Fish and Shellfish Sampled in the Ocean Area Within 20Km Radius of Fukushima Daiichi NPS* (Sampling Period: January – March, 2014)
Investigation Report of Fish and Shellfish Sampled in the Ocean Area Within 20km Radius of Fukushima Daiichi NPS* (Sampling period: January – March, 2014) Tokyo Electric Power Company June 13, 2014 * Exclude the data obtained in the port of Fukushima Daiichi NPS 1. Purpose of the Investigation of Fish and Shellfish Sampled in the Ocean Area Within 20km Radius of Fukushima Daiichi NPS (1) To understand radioactive cesium density by fish species - Comparison with the food standard value (total cesium amount: 100Bq/kg) (2) To understand the geographical distribution of radioactive cesium density of fish and shellfish - Sampling at fixed measurement points (gill net fishing, trawl net fishing) (3) To understand the change of radioactive cesium density of fish and shellfish over time - Accumulating basic data in order to forecast trends 1 2-1. Investigation Results (Radioactive Cesium Density by Fish Species) Approx. 90% of all the measurement results were below the standard value. Standard value: 100 Bq/kg of total amount of radioactive cesium Sampling period: January to March , 2014 Sampling period: October to November , 2013 [Top 3 Density Levels] [Top 3 Density Levels] Number of fish 32 (Unit: Bq/kg (Raw)) 39 (Unit: Bq/kg (Raw)) species (cesium 1. Common skete (cesium exceeding 1. Banded houndshark exceeding the 2. Schlegel's black rockfish the 2. Schlegel's black rockfish standard value: 3. Sebastes cheni standard value: 9) 3. Marbled sole ) 6 [Samples below the [Samples below the detection limit] detection limit] 1. Blue crab 1. Blue crab Number of 241 2. Yellow goosefish 271 2. Roundnose flounder measurements (cesium 3. -

Elasmobranch Biodiversity, Conservation and Management Proceedings of the International Seminar and Workshop, Sabah, Malaysia, July 1997
The IUCN Species Survival Commission Elasmobranch Biodiversity, Conservation and Management Proceedings of the International Seminar and Workshop, Sabah, Malaysia, July 1997 Edited by Sarah L. Fowler, Tim M. Reed and Frances A. Dipper Occasional Paper of the IUCN Species Survival Commission No. 25 IUCN The World Conservation Union Donors to the SSC Conservation Communications Programme and Elasmobranch Biodiversity, Conservation and Management: Proceedings of the International Seminar and Workshop, Sabah, Malaysia, July 1997 The IUCN/Species Survival Commission is committed to communicate important species conservation information to natural resource managers, decision-makers and others whose actions affect the conservation of biodiversity. The SSC's Action Plans, Occasional Papers, newsletter Species and other publications are supported by a wide variety of generous donors including: The Sultanate of Oman established the Peter Scott IUCN/SSC Action Plan Fund in 1990. The Fund supports Action Plan development and implementation. To date, more than 80 grants have been made from the Fund to SSC Specialist Groups. The SSC is grateful to the Sultanate of Oman for its confidence in and support for species conservation worldwide. The Council of Agriculture (COA), Taiwan has awarded major grants to the SSC's Wildlife Trade Programme and Conservation Communications Programme. This support has enabled SSC to continue its valuable technical advisory service to the Parties to CITES as well as to the larger global conservation community. Among other responsibilities, the COA is in charge of matters concerning the designation and management of nature reserves, conservation of wildlife and their habitats, conservation of natural landscapes, coordination of law enforcement efforts as well as promotion of conservation education, research and international cooperation. -

An Unspotted, Grey, Rather Stocky Mustelus with Strongly Cuspidate Teeth, Lanceolate Denticles, Short Caudal Peduncle, and Broadly Frayed Posterior Dorsal Fin Margins
click for previous page - 426 - Field Marks: An unspotted, grey, rather stocky Mustelus with strongly cuspidate teeth, lanceolate denticles, short caudal peduncle, and broadly frayed posterior dorsal fin margins. Diagnostic Features : Body fairly stocky, almost humpbacked. Head fairly long, prepectoral length 20 to 24% of total length; snout moderately long and bluntly angular in lateral view, preoral snout 5.6 to 7.6% of total length, preorbital snout 6.7 to 8.5% of total length; internarial space very broad, 2.9 to 3.7% of total length; eyes fairly large, eye length .2 to 3.1 times in preorbital snout and 2.1 to 3.2% of total length; interorbital space moderately broad, 4.5 to 5.6% of total length; mouth moderately long, slightly longer than eye length and 2.6 to 3.7% of total length; upper labial furrows considerably longer than lowers and 2.6 to 3.7% of total length; teeth cuspidate and asymmetric, with a prominent primary cusp and low cusplets occasionally present; buccopharyn- geal denticles confined to anterior fourth of palate and tongue tip. Interdorsal space 16 to 21% of total length; trailing edges of dorsal fins naked, with a conspicuous dark margin of bare ceratotrichia; first dorsal broadly triangular, with posteroventrally sloping posterior margin, midbase closer to pelvic bases than pectorals; pectoral fins fairly large, length of anterior margins 14 to 17% of total length, width of posterior margins 11 to 14% of total length; pelvic fins moderately large, length of anterior margins 7.1 to 9.4% of total length; anal fin height 2.3 to 3.8% of total length; anal-caudal space less than or subequal to second dorsal height, and 4.7 to 7.4% of total length; ventral caudal lobe hardly falcate in adults. -
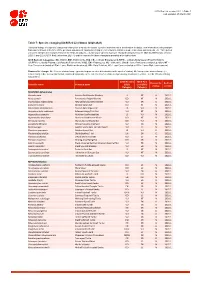
Table 7: Species Changing IUCN Red List Status (2020-2021)
IUCN Red List version 2021-1: Table 7 Last Updated: 25 March 2021 Table 7: Species changing IUCN Red List Status (2020-2021) Published listings of a species' status may change for a variety of reasons (genuine improvement or deterioration in status; new information being available that was not known at the time of the previous assessment; taxonomic changes; corrections to mistakes made in previous assessments, etc. To help Red List users interpret the changes between the Red List updates, a summary of species that have changed category between 2020 (IUCN Red List version 2020-3) and 2021 (IUCN Red List version 2021-1) and the reasons for these changes is provided in the table below. IUCN Red List Categories: EX - Extinct, EW - Extinct in the Wild, CR - Critically Endangered [CR(PE) - Critically Endangered (Possibly Extinct), CR(PEW) - Critically Endangered (Possibly Extinct in the Wild)], EN - Endangered, VU - Vulnerable, LR/cd - Lower Risk/conservation dependent, NT - Near Threatened (includes LR/nt - Lower Risk/near threatened), DD - Data Deficient, LC - Least Concern (includes LR/lc - Lower Risk, least concern). Reasons for change: G - Genuine status change (genuine improvement or deterioration in the species' status); N - Non-genuine status change (i.e., status changes due to new information, improved knowledge of the criteria, incorrect data used previously, taxonomic revision, etc.); E - Previous listing was an Error. IUCN Red List IUCN Red Reason for Red List Scientific name Common name (2020) List (2021) change version Category -

ASFIS ISSCAAP Fish List February 2007 Sorted on Scientific Name
ASFIS ISSCAAP Fish List Sorted on Scientific Name February 2007 Scientific name English Name French name Spanish Name Code Abalistes stellaris (Bloch & Schneider 1801) Starry triggerfish AJS Abbottina rivularis (Basilewsky 1855) Chinese false gudgeon ABB Ablabys binotatus (Peters 1855) Redskinfish ABW Ablennes hians (Valenciennes 1846) Flat needlefish Orphie plate Agujón sable BAF Aborichthys elongatus Hora 1921 ABE Abralia andamanika Goodrich 1898 BLK Abralia veranyi (Rüppell 1844) Verany's enope squid Encornet de Verany Enoploluria de Verany BLJ Abraliopsis pfefferi (Verany 1837) Pfeffer's enope squid Encornet de Pfeffer Enoploluria de Pfeffer BJF Abramis brama (Linnaeus 1758) Freshwater bream Brème d'eau douce Brema común FBM Abramis spp Freshwater breams nei Brèmes d'eau douce nca Bremas nep FBR Abramites eques (Steindachner 1878) ABQ Abudefduf luridus (Cuvier 1830) Canary damsel AUU Abudefduf saxatilis (Linnaeus 1758) Sergeant-major ABU Abyssobrotula galatheae Nielsen 1977 OAG Abyssocottus elochini Taliev 1955 AEZ Abythites lepidogenys (Smith & Radcliffe 1913) AHD Acanella spp Branched bamboo coral KQL Acanthacaris caeca (A. Milne Edwards 1881) Atlantic deep-sea lobster Langoustine arganelle Cigala de fondo NTK Acanthacaris tenuimana Bate 1888 Prickly deep-sea lobster Langoustine spinuleuse Cigala raspa NHI Acanthalburnus microlepis (De Filippi 1861) Blackbrow bleak AHL Acanthaphritis barbata (Okamura & Kishida 1963) NHT Acantharchus pomotis (Baird 1855) Mud sunfish AKP Acanthaxius caespitosa (Squires 1979) Deepwater mud lobster Langouste -

Results of Radioactive Material Monitoring of Aquatic Organisms (Location G Off Iwaki City (Nakoso))
○ Results of Radioactive Material Monitoring of Aquatic Organisms (Location G off Iwaki City (Nakoso)) <Location G off Iwaki City (Nakoso): Samples collected> Items General items Radioactive materials Locations Water Sediment water (Cs) water (Sr) Sediment (Cs) Sediment (Sr) G-1 ○ ○ ○ ○ ○ ○ G-2 - ○ - - ○ - G-3 - ○ - - ○ - G-4 ○ ○ ○ ○ ○ ○ G-5 - ○ - - ○ - <Location G off Iwaki City (Nakoso) Site measurement item> Latitude and longitude of the Water Sediment Other Items location Survey date and time Latitude Longitude Water temperature Sediment temperature Property Color Odor Contaminants Secchi disk depth Water depth Locations G-1 8:25 36.86550 140.89238 10.1 9.1 Sand 2.5Y3/2 None None 10.6 38.8 G-2 9:20 36.90053 140.92147 - 9.3 Sand 2.5Y3/1 None None - 35.1 G-3 2012/1/13 9:42 36.91210 140.94050 - 9.3 Sand 2.5Y2/2 None None - 33.8 G-4 10:11 36.91855 140.96768 11.0 9.8 Sand 2.5Y2/2 None None 10.6 36.0 G-5 10:50 36.92990 140.99013 - 9.1 Sand 2.5Y2/2 None Shellfish fry ― 44.9 <Location G off Iwaki City (Nakoso): Analysis items Water> Latitude and longitude of the Electrical pH BOD COD DO Salinity TOC SS Turbidity Cs-134 Cs-137 Sr-90 Items Survey date and time location conductivity (Bq/L) (Bq/L) (Bq/L) Locations Latitude Longitude - (mg/L) (mg/L) (mg/L) (mS/m) - (mg/L) (mg/L) (FNU) G-1 8:25 36.86550 140.89238 8.1 0.7 0.8 9.0 4,990 33.97 0.7 4 0.8 0.039 0.050 0.011 G-2 9:20 36.90053 140.92147 - - - - - - - - - - - - G-3 2012/1/13 9:42 36.91210 140.94050 - - - - - - - - - - - - G-4 10:11 36.91855 140.96768 8.1 <0.5 0.7 9.0 4,990 33.96 0.6 1 0.6 -

And Their Functional, Ecological, and Evolutionary Implications
DePaul University Via Sapientiae College of Science and Health Theses and Dissertations College of Science and Health Spring 6-14-2019 Body Forms in Sharks (Chondrichthyes: Elasmobranchii), and Their Functional, Ecological, and Evolutionary Implications Phillip C. Sternes DePaul University, [email protected] Follow this and additional works at: https://via.library.depaul.edu/csh_etd Part of the Biology Commons Recommended Citation Sternes, Phillip C., "Body Forms in Sharks (Chondrichthyes: Elasmobranchii), and Their Functional, Ecological, and Evolutionary Implications" (2019). College of Science and Health Theses and Dissertations. 327. https://via.library.depaul.edu/csh_etd/327 This Thesis is brought to you for free and open access by the College of Science and Health at Via Sapientiae. It has been accepted for inclusion in College of Science and Health Theses and Dissertations by an authorized administrator of Via Sapientiae. For more information, please contact [email protected]. Body Forms in Sharks (Chondrichthyes: Elasmobranchii), and Their Functional, Ecological, and Evolutionary Implications A Thesis Presented in Partial Fulfilment of the Requirements for the Degree of Master of Science June 2019 By Phillip C. Sternes Department of Biological Sciences College of Science and Health DePaul University Chicago, Illinois Table of Contents Table of Contents.............................................................................................................................ii List of Tables..................................................................................................................................iv -

Triakis Scyllium
www.nature.com/scientificreports OPEN Cross‑sectional anatomy, computed tomography, and magnetic resonance imaging of the banded houndshark (Triakis scyllium) Sang Wha Kim 1,5, Adams Hei Long Yuen 1,5, Cherry Tsz Ching Poon 2, Joon Oh Hwang3, Chang Jun Lee3, Moon‑Kwan Oh4, Ki Tae Kim1, Hyoun Joong Kim1, Sib Sankar Giri1, Sang Guen Kim1, Jun Kwon1, Sung Bin Lee1, Min Cheol Choi1 & Se Chang Park1* Due to their important phylogenetic position among extant vertebrates, sharks are an invaluable group in evolutionary developmental biology studies. A thorough understanding of shark anatomy is essential to facilitate these studies and documentation of this iconic taxon. With the increasing availability of cross‑sectional imaging techniques, the complicated anatomy of both cartilaginous and soft tissues can be analyzed non‑invasively, quickly, and accurately. The aim of this study is to provide a detailed anatomical description of the normal banded houndshark (Triakis scyllium) using computed tomography (CT) and magnetic resonance imaging (MRI) along with cryosection images. Three banded houndsharks were scanned using a 64‑detector row spiral CT scanner and a 3 T MRI scanner. All images were digitally stored and assessed using open‑source Digital Imaging and Communications in Medicine viewer software in the transverse, sagittal, and dorsal dimensions. The banded houndshark cadavers were then cryosectioned at approximately 1‑cm intervals. Corresponding transverse cryosection images were chosen to identify the best anatomical correlations for transverse CT and MRI images. The resulting images provided excellent detail of the major anatomical structures of the banded houndshark. The illustrations in the present study could be considered as a useful reference for interpretation of normal and pathological imaging studies of sharks. -

Survey Report for Fish and Seashells of the Sea Within a 20 Km Radius Of
Survey report* for fish and seashells of the sea within a 20 km radius of Fukushima Daiichi Nuclear Power Station (Samples taken during the period between July and September, 2014) December 5, 2014 Tokyo Electric Power Company * Except for the survey data obtained inside the port of Fukushima Daiichi Nuclear Power Station. 1. Objectives for surveying fish and seashells of the sea within a 20 km radius of Fukushima Daiichi NPS (1) Grasp radioactive cesium levels of each fish species. ・Comparison with the food product standards (the total of cesium: 100 Bq/ kg) (2) Grasp the areal distribution of radioactive cesium levels in fish and seashells. ・Sampling at fixed surveying spots (gillnet and trawler fisheries) (3) Grasp the transition with time of radioactive cesium levels of fish and seashells. ・Basic data taken for the transition prediction. 1 2‐1. Survey results (radioactive cesium levels by fish species) More than 99% of the samples taken indicate that the measured levels fall below the standard value. Standard value: a total of 100 radioactive cesium (Bq/kg) Samples taken during Jul. to Sep. 2014. Samples taken during Apr. to Jun. 2014. [Top 3 species with high levels] [Top 3 species with high levels] (Unit: Bq/kg) (Unit: Bq/kg) 38 1) Common skete 131 42 1) Common skete 370 2) Stone flounder 96 2) Sebastes chen 199 No. of species (of which, 1 case (of which, 4 cases 3) Microstomus achne 91 3) Microstomus achne 176 exceeded the exceeded the standard value) standard value) [Below the detection limit value] [Below the detection limit value] 1) Zenopsis nebulosa, 1) crimson sea bream 2) Yellow goosefish, 2) White croaker 3) Japanese amberjack, 3) Banded houndshark No. -

Nurse Shark (Ginglymostoma Cirratum) BAC Library
Proposal for construction of a nurse shark (Ginglymostoma cirratum) BAC library Martin Flajnik and Yuko Ohta, Dept. of Microbiology and Immunology, University of Maryland School of Medicine, Baltimore, MD 21201 1. The importance of the nurse shark to biomedical or biological research The cartilaginous fish, specifically the elasmobranchs (sharks, skates, and rays) are the oldest group of living vertebrates shown to have developed an adaptive immune system with underlying molecules and mechanisms similar to those of mammals. This ancient taxon is the oldest with jaws and diverged from the common ancestor of all vertebrates 460-520 million years ago (the oldest group of jawed vertebrates, the placoderms, are extinct). A second evolutionary wave occurred about 250 million years ago, which gave rise to extant elasmobranchs (1). Studies of this old group, in comparison to representatives in other vertebrate taxa, allow us to theorize about fundamental genetic, developmental, and functional characteristics in the common ancestor of all vertebrates. Our work focuses on evolution of the adaptive immune system (2), and our aim is to uncover those essential elements in all immune systems as well as to perhaps reveal some characteristics that were evolutionary forerunners of the human system (the former task obviously much easier than the latter!) The comparative approach has been instrumental in deciphering the necessary characteristics of the adaptive immune system (3). Among elasmobranch species, we and others use the nurse shark Ginglymostoma cirratum to study the genetics, biochemistry, and mechanisms that regulate the immune system. Compared to other shark species, the nurse shark genome size is relatively small, only slightly larger than that of humans.