Convergent Evolution of Morphological and Ecological Traits
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Disaggregation of Bird Families Listed on Cms Appendix Ii
Convention on the Conservation of Migratory Species of Wild Animals 2nd Meeting of the Sessional Committee of the CMS Scientific Council (ScC-SC2) Bonn, Germany, 10 – 14 July 2017 UNEP/CMS/ScC-SC2/Inf.3 DISAGGREGATION OF BIRD FAMILIES LISTED ON CMS APPENDIX II (Prepared by the Appointed Councillors for Birds) Summary: The first meeting of the Sessional Committee of the Scientific Council identified the adoption of a new standard reference for avian taxonomy as an opportunity to disaggregate the higher-level taxa listed on Appendix II and to identify those that are considered to be migratory species and that have an unfavourable conservation status. The current paper presents an initial analysis of the higher-level disaggregation using the Handbook of the Birds of the World/BirdLife International Illustrated Checklist of the Birds of the World Volumes 1 and 2 taxonomy, and identifies the challenges in completing the analysis to identify all of the migratory species and the corresponding Range States. The document has been prepared by the COP Appointed Scientific Councilors for Birds. This is a supplementary paper to COP document UNEP/CMS/COP12/Doc.25.3 on Taxonomy and Nomenclature UNEP/CMS/ScC-Sc2/Inf.3 DISAGGREGATION OF BIRD FAMILIES LISTED ON CMS APPENDIX II 1. Through Resolution 11.19, the Conference of Parties adopted as the standard reference for bird taxonomy and nomenclature for Non-Passerine species the Handbook of the Birds of the World/BirdLife International Illustrated Checklist of the Birds of the World, Volume 1: Non-Passerines, by Josep del Hoyo and Nigel J. Collar (2014); 2. -

Bird Watching in Cyprus a Brief Guide for Visitors To
BIRD WATCHING IN CYPRUS A BRIEF GUIDE FOR VISITORS TO THE ISLAND 1 Information on Cyprus in general The position of Cyprus in the eastern Mediterranean with Turkey to the north, Syria to the east and Egypt to the south, places it on one of the major migration routes in the Mediterranean and makes it a stop off point for many species which pass each year from Europe/Asia to Africa via the Nile Delta. The birds that occur regularly on passage form a large percentage of the ‘Cyprus list’ that currently totals nearly 380 species. Of these only around 50 are resident and around 40 are migrant species that regularly or occasionally breed. The number of birds passing over during the spring and autumn migration periods are impressive, as literally millions of birds pour through Cyprus. Spring migration gets underway in earnest around the middle of March, usually depending on how settled the weather is, and continues into May. A few early arrivals can even be noted in February, especially the swallows, martins and swifts, some wheatears and the Great Spotted Cuckoo Clamator glandarius. Slender-billed Gulls Larus genei and herons can be seen in flocks along the coastline. Each week seems to provide a different species to watch for. The end of March sees Roller Coracias garrulous, Masked Shrike Lanius nubicus, Cretzschmar’s Bunting Emberiza caesia, Black-headed Wagtails Motacilla flava feldegg and Red-rumped Swallows Cecropsis daurica, while on the wetlands Marsh Sandpipers Tringa stagnatilis, Collared Pratincole Glareola pratincola, Spur-winged Vanellus spinosus and Greater Sand Plover Charadrius leschenaultii can be seen. -

Territory Size of the Mourning Wheatear Oenanthe Lugens Along an Aridity Gradient
Journal of Arid Environments 74 (2010) 1413e1417 Contents lists available at ScienceDirect Journal of Arid Environments journal homepage: www.elsevier.com/locate/jaridenv Territory size of the Mourning Wheatear Oenanthe lugens along an aridity gradient F. Khoury a,*, N. Boulad b a Department of Biological Sciences and Biotechnology, The Hashemite University, P.O. Box 150459, Zarqa 13115, Jordan b Research and Survey Section, Royal Society for the Conservation of Nature, P.O.Box 1215, Jubeiha 11941, Amman, Jordan article info abstract Article history: Breeding territories of Mourning Wheatear Oenanthe lugens were investigated in three study areas in Received 15 September 2009 Jordan to describe and explain variation in territory size along an aridity gradient. In moderately Received in revised form productive semi-deserts, Mourning Wheatears defended small territories (averages for two different 22 January 2010 areas 3.0 ha and 5.9 ha) that were arranged as relatively dense neighbourhood clusters, while in Accepted 11 May 2010 extremely arid deserts, territories were significantly larger (average 22.8 ha) and arranged in very loose Available online 9 June 2010 clusters. Contender pressure and probably vegetation cover (as cue of sustained prey availability) played a dual role in determining territory sizes. The relative role of prey availability as proximate factor Keywords: Contender pressure increased with aridity due to low contender pressure in extremely arid deserts. Ó Desert 2010 Elsevier Ltd. All rights reserved. Jordan Productivity Territoriality 1. Introduction Wheatears, genus Oenanthe, are insectivorous birds that aggressively defend exclusive, food-related breeding territories Territory size in animals often relates inversely to food abun- against both conspecific intruders and heterospecific competitors dance (food-value theory), i.e. -

Southern Israel: a Spring Migration Spectacular
SOUTHERN ISRAEL: A SPRING MIGRATION SPECTACULAR MARCH 21–APRIL 3, 2019 Spectacular male Bluethroat (orange spotted form) in one of the world’s greatest migration hotspots, Eilat © Andrew Whittaker LEADERS: ANDREW WHITTAKER & MEIDAD GOREN LIST COMPILED BY: ANDREW WHITTAKER VICTOR EMANUEL NATURE TOURS, INC. 2525 WALLINGWOOD DRIVE, SUITE 1003 AUSTIN, TEXAS 78746 WWW.VENTBIRD.COM SOUTHERN ISRAEL: A SPRING MIGRATION SPECTACULAR March 21–April 3, 2019 By Andrew Whittaker The sky was full of migrating White Storks in the thousands above Masada and parts of the the Negev Desert © Andrew Whittaker My return to Israel after working in Eilat banding birds some 36 years ago certainly was an exciting prospect and a true delight to witness, once again, one of the world’s most amazing natural phenomena, avian migration en masse. This delightful tiny country is rightly world-renowned as being the top migration hotspot, with a staggering estimated 500–750 million birds streaming through the African- Eurasian Flyway each spring, comprising over 200 different species! Israel is truly an unparalleled destination allowing one to enjoy this exceptional spectacle, especially in the spring when all are in such snazzy breeding plumage. Following the famous Great Rift Valley that bisects Israel, they migrate thousands of miles northwards from their wintering grounds in western Africa bound for rich breeding grounds, principally in central and eastern Europe. Israel acts as an amazing bottleneck resulting in an avian abundance everywhere you look: skies filled with countless migratory birds from storks to raptors; Victor Emanuel Nature Tours 2 Southern Israel, 2019 rich fish ponds and salt flats holding throngs of flamingos, shorebirds, and more; and captivating deserts home to magical regional goodies such as sandgrouse, bustards and larks, while every bush and tree are moving with warblers. -

Vickery Et Al. 2014, Cresswell 2014)
1 Accepted Journal of Avian Biology 25/8/16 MS JAV-1119 R2 2 Cyprus Wheatears Oenanthe cypriaca likely reach sub-Saharan African wintering 3 grounds in a single migratory flight 4 5 Marina Xenophontos, Emma Blackburn & Will Cresswell* 6 Centre for Biological Diversity, University of St Andrews, Harold Mitchell Building, St Andrews, Fife KY16 7 9TH, UK 8 9 *Correspondence author: [email protected] 10 11 1 12 Long-distance migratory flights with multiple stop-overs, multiple wintering sites, and small-scale 13 connectivity in Afro-Palearctic migrants are likely to increase their vulnerability to environmental change 14 and lead to declining populations. Here we present the migration tracks and wintering locations of the first 15 six Cyprus Wheatears to be tracked with geolocators: a species with high survival and a stable 16 population. We therefore predicted a non-stop flight from Cyprus to sub-Saharan wintering grounds, a 17 single wintering area for each individual and a wide spread of wintering locations representing low 18 migratory connectivity at the population level. The sub-Saharan wintering grounds in South Sudan, Sudan 19 and Ethiopia were likely reached by a single flight of an average straight-line distance of 2,538 km in ca. 20 60 hours, with an average minimum speed of 43.1 km/h. The high speed of migration probably ruled out 21 stop-overs greater than a few hours. Cyprus Wheatears migrated from Cyprus in mid-late October and 22 most probably remained at a single location throughout winter; three out of five birds with available data 23 may have used a second site <100 km away during February; all returned between the 7 – 22nd March 24 when accurate geolocation data are not possible due to the equinox. -

Bird Survey of South-Eastern Laikipia: Lolldaiga Ranch, Ole Naishu Ranch, Borana Ranch, and Mukogodo Forest Reserve
8 November 2015 Dear All, Recently Nigel Hunter and I went to stay with Tom Butynski on Lolldaiga Hills Ranch. Whilst there we were joined by Paul Benson, and Eleanor Monbiot for the 31st Oct, Chris Thouless joined us on 1st Nov in Mukogodo, and he and Caroline kindly put the three of us up at their house for the nights of 31st Oct and 1st Nov., and for both these dates we enjoyed the company of Lawrence, the bird-guide at Borana Lodge. For our full day on Lolldaiga on 2nd Nov., Paul spent the entire day with us. The more interesting observations follow, but this is far from the full list which exceeded 200 on Lolldaiga alone in spite of the relatively short time we were there. Best for now Brian BIRD SURVEY OF SOUTH-EASTERN LAIKIPIA: LOLLDAIGA RANCH, OLE NAISHU RANCH, BORANA RANCH, AND MUKOGODO FOREST RESERVE ITINERARY 30th Oct 2015 Drove Nairobi to Lolldaiga, birded as far as old Maize Paddock in late afternoon. 31st Oct Drove from TB house out through Ole Naishu Ranch and across Borana arriving at Mukogodo Forest in early afternoon. 1st Nov All day in Mukogodo Forest, and just 5 kilometres down the main descent road in afternoon. 2nd Nov All day on Borana, back across Ole Naishu to Lolldaiga. 3rd Nov All day outing on Lolldaiga to Black Rock, Ngainitu Kopje (North Gate), Sinyai Lugga, and evening near the Monument. 4th Nov Morning on descent road to Main Gate, Lolldaiga and forest along Timau River, leaving 11.15 AM for Nairobi. -

Ethiopian Endemics I 11Th to 29Th January 2014 & Lalibela Historical Extension 29Th January to 1St February 2014
Ethiopian Endemics I 11th to 29th January 2014 & Lalibela Historical Extension th st 29 January to 1 February 2014 Trip report Abyssinian Roller by Markus Lilje Tour leaders: Wayne Jones & Andrew Stainthorpe. Trip report compiled by Wayne Jones RBT Ethiopian Endemics I Trip Report 2014 2 Top 10 birds as voted by participants: 1. Ruspoli’s Turaco 2. Abyssinian Roller 3. Half-collared Kingfisher 4. Fox Kestrel 5. Abyssinian Ground Thrush 6. Nile Valley Sunbird 7. Hartlaub’s Bustard 8. Quailfinch 9. Abyssinian Catbird 10. Abyssinian Woodpecker Tour Summary Our tour kicked off in the grounds of our hotel in Addis Ababa on what was, essentially, an arrival day. Despite its location in the middle of the bustling and chaotic capital city, the gardens yielded a good selection of birds including Wattled Ibis, African Harrier-Hawk, White-collared Pigeon, African Paradise Flycatcher, Brown Parisoma, Dusky Turtle Dove, Abyssinian Thrush, Montane White-eye, Abyssinian Slaty Flycatcher, Brown-rumped Seedeater and Ruppell’s Robin-Chat. Common Cranes by Adam Riley We set out early the following morning so as to arrive at Lake Chelekcheka just after dawn, when the hundreds of Common Cranes that roost there start becoming active amid a cacophony of guttural bugling. With waves of cranes passing over us on their way to forage in the fields, we found plenty of other waterbirds including Northern Shoveler, Spur-winged Goose, Northern Pintail, Eurasian Teal, Greater and Lesser Flamingos, Spur-winged Lapwing, Three-banded Plover, Black-tailed Godwit and Temminck’s Stint. Yellow Wagtails abounded and one of the area’s specials, the tiny and gorgeous Quailfinch, gave excellent views. -

South Africa Mega Birding III 5Th to 27Th October 2019 (23 Days) Trip Report
South Africa Mega Birding III 5th to 27th October 2019 (23 days) Trip Report The near-endemic Gorgeous Bushshrike by Daniel Keith Danckwerts Tour leader: Daniel Keith Danckwerts Trip Report – RBT South Africa – Mega Birding III 2019 2 Tour Summary South Africa supports the highest number of endemic species of any African country and is therefore of obvious appeal to birders. This South Africa mega tour covered virtually the entire country in little over a month – amounting to an estimated 10 000km – and targeted every single endemic and near-endemic species! We were successful in finding virtually all of the targets and some of our highlights included a pair of mythical Hottentot Buttonquails, the critically endangered Rudd’s Lark, both Cape, and Drakensburg Rockjumpers, Orange-breasted Sunbird, Pink-throated Twinspot, Southern Tchagra, the scarce Knysna Woodpecker, both Northern and Southern Black Korhaans, and Bush Blackcap. We additionally enjoyed better-than-ever sightings of the tricky Barratt’s Warbler, aptly named Gorgeous Bushshrike, Crested Guineafowl, and Eastern Nicator to just name a few. Any trip to South Africa would be incomplete without mammals and our tally of 60 species included such difficult animals as the Aardvark, Aardwolf, Southern African Hedgehog, Bat-eared Fox, Smith’s Red Rock Hare and both Sable and Roan Antelopes. This really was a trip like no other! ____________________________________________________________________________________ Tour in Detail Our first full day of the tour began with a short walk through the gardens of our quaint guesthouse in Johannesburg. Here we enjoyed sightings of the delightful Red-headed Finch, small numbers of Southern Red Bishops including several males that were busy moulting into their summer breeding plumage, the near-endemic Karoo Thrush, Cape White-eye, Grey-headed Gull, Hadada Ibis, Southern Masked Weaver, Speckled Mousebird, African Palm Swift and the Laughing, Ring-necked and Red-eyed Doves. -
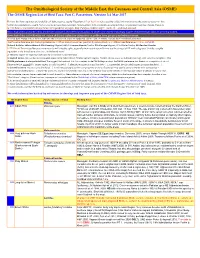
OSME List V3.4 Passerines-2
The Ornithological Society of the Middle East, the Caucasus and Central Asia (OSME) The OSME Region List of Bird Taxa: Part C, Passerines. Version 3.4 Mar 2017 For taxa that have unproven and probably unlikely presence, see the Hypothetical List. Red font indicates either added information since the previous version or that further documentation is sought. Not all synonyms have been examined. Serial numbers (SN) are merely an administrative conveninence and may change. Please do not cite them as row numbers in any formal correspondence or papers. Key: Compass cardinals (eg N = north, SE = southeast) are used. Rows shaded thus and with yellow text denote summaries of problem taxon groups in which some closely-related taxa may be of indeterminate status or are being studied. Rows shaded thus and with white text contain additional explanatory information on problem taxon groups as and when necessary. A broad dark orange line, as below, indicates the last taxon in a new or suggested species split, or where sspp are best considered separately. The Passerine Reference List (including References for Hypothetical passerines [see Part E] and explanations of Abbreviated References) follows at Part D. Notes↓ & Status abbreviations→ BM=Breeding Migrant, SB/SV=Summer Breeder/Visitor, PM=Passage Migrant, WV=Winter Visitor, RB=Resident Breeder 1. PT=Parent Taxon (used because many records will antedate splits, especially from recent research) – we use the concept of PT with a degree of latitude, roughly equivalent to the formal term sensu lato , ‘in the broad sense’. 2. The term 'report' or ‘reported’ indicates the occurrence is unconfirmed. -

Protected Area Management Plan Development - SAPO NATIONAL PARK
Technical Assistance Report Protected Area Management Plan Development - SAPO NATIONAL PARK - Sapo National Park -Vision Statement By the year 2010, a fully restored biodiversity, and well-maintained, properly managed Sapo National Park, with increased public understanding and acceptance, and improved quality of life in communities surrounding the Park. A Cooperative Accomplishment of USDA Forest Service, Forestry Development Authority and Conservation International Steve Anderson and Dennis Gordon- USDA Forest Service May 29, 2005 to June 17, 2005 - 1 - USDA Forest Service, Forestry Development Authority and Conservation International Protected Area Development Management Plan Development Technical Assistance Report Steve Anderson and Dennis Gordon 17 June 2005 Goal Provide support to the FDA, CI and FFI to review and update the Sapo NP management plan, establish a management plan template, develop a program of activities for implementing the plan, and train FDA staff in developing future management plans. Summary Week 1 – Arrived in Monrovia on 29 May and met with Forestry Development Authority (FDA) staff and our two counterpart hosts, Theo Freeman and Morris Kamara, heads of the Wildlife Conservation and Protected Area Management and Protected Area Management respectively. We decided to concentrate on the immediate implementation needs for Sapo NP rather than a revision of existing management plan. The four of us, along with Tyler Christie of Conservation International (CI), worked in the CI office on the following topics: FDA Immediate -

EUROPEAN BIRDS of CONSERVATION CONCERN Populations, Trends and National Responsibilities
EUROPEAN BIRDS OF CONSERVATION CONCERN Populations, trends and national responsibilities COMPILED BY ANNA STANEVA AND IAN BURFIELD WITH SPONSORSHIP FROM CONTENTS Introduction 4 86 ITALY References 9 89 KOSOVO ALBANIA 10 92 LATVIA ANDORRA 14 95 LIECHTENSTEIN ARMENIA 16 97 LITHUANIA AUSTRIA 19 100 LUXEMBOURG AZERBAIJAN 22 102 MACEDONIA BELARUS 26 105 MALTA BELGIUM 29 107 MOLDOVA BOSNIA AND HERZEGOVINA 32 110 MONTENEGRO BULGARIA 35 113 NETHERLANDS CROATIA 39 116 NORWAY CYPRUS 42 119 POLAND CZECH REPUBLIC 45 122 PORTUGAL DENMARK 48 125 ROMANIA ESTONIA 51 128 RUSSIA BirdLife Europe and Central Asia is a partnership of 48 national conservation organisations and a leader in bird conservation. Our unique local to global FAROE ISLANDS DENMARK 54 132 SERBIA approach enables us to deliver high impact and long term conservation for the beneit of nature and people. BirdLife Europe and Central Asia is one of FINLAND 56 135 SLOVAKIA the six regional secretariats that compose BirdLife International. Based in Brus- sels, it supports the European and Central Asian Partnership and is present FRANCE 60 138 SLOVENIA in 47 countries including all EU Member States. With more than 4,100 staf in Europe, two million members and tens of thousands of skilled volunteers, GEORGIA 64 141 SPAIN BirdLife Europe and Central Asia, together with its national partners, owns or manages more than 6,000 nature sites totaling 320,000 hectares. GERMANY 67 145 SWEDEN GIBRALTAR UNITED KINGDOM 71 148 SWITZERLAND GREECE 72 151 TURKEY GREENLAND DENMARK 76 155 UKRAINE HUNGARY 78 159 UNITED KINGDOM ICELAND 81 162 European population sizes and trends STICHTING BIRDLIFE EUROPE GRATEFULLY ACKNOWLEDGES FINANCIAL SUPPORT FROM THE EUROPEAN COMMISSION. -

Wheatears of the Oenanthe Lugens Complex
around our heads and calling, and occasionally This is the first breeding record of European perching on rocks allowing scrutiny through a roller for Arabia. More conclusive breeding telescope. There were two or more females, one evidence will be sought in 1991 when their imF.ature male and one adult male, showing the presence at Digdaga will be monitored more diagnostic bluish hood and unspotted bright closely. chestnut mantle. Some of the birds also had projecting central tail feathers (or perhaps just Colin Richardson, P.O. Box 2825, Dubai, UAE. lack of white tips to these feathers). The juvenile was disturbed from a rock ledge, but was NB The European roller breeds throughout much of not yet able to fly. Iran, in Iraq at the head of the Arabian Gulf and probably in Jordan. Ed. A month later, on 3 July, another adult male was seen 50 km or so to the south, but apart from WHEATEARS OF THE OENANTHE LUGENS COMPLEX these records and another sighting of a probable (MOURNING WHEATEAR) IN ARABIA female in spring 1989, records of lesser kestrels in the region seem to be very few and far Mike Jennings recently (1989, Phoenix 6) between. Green (1984, Sandgrouse 6:48-50) makes expressed the view that South Arabian mourning no mention of the species. However, on the same wheatears Oenanthe "Iugentoides" should be mountain in March 1988, a group of kestrels seen considered a separate species from O.Iugens. from a distance by Paul Goriup and Peter Symens Hollom et al. (Birds of the Middle East and were possibly this species.