Question Bank
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mcb 407-Immunology and Immunochemistry-Lecture Note
MCB 407-IMMUNOLOGY AND IMMUNOCHEMISTRY-LECTURE NOTE DR. D. A. OJO BRIEF HISTORICAL REVIEW OF IMMUNOLOGY The mechanism by which antibody are formed has been debated for years. It was proposed that the specificity of an antibody molecule was determined both by its amino acid sequence but by the molding of the peptide chain around the antigenic determinant. This theory lost favour when it became apparent that antibody-forming cells were devoid of antigen and that antibody specificity was a function of amino acid sequence. At present, the CLONAL (proposed by Burnete) SELECTION THEORY is widely accepted. It holds that an immunologically responsive cell can respond to only one antigen or a closely related group of antigens and that this property is inherent in the cell before the antigen is encountered. According to the clonal selection theory, each individual is endowed with a very large pool of lymphocytes, each of which is capable of responding to a different antigen. When the antigen enters the body, it selects the lymphocyte which has the best “fit” by virtue of a surface receptor. The antigen binds to this antibody-like receptor, and the cell is stimulated to proliferate and form a clone of cells. Thus, selected cells quickly differentiate into plasma cells and secrete antibody which is specific for the antigen which served as the original selecting agent (or a closely related group of antigens). The History of Blood Transfusion Man’s centuries-long desire to perform blood transfusion as a therapeutic procedure forms the cornerstone of the modern science of immunohematology. At present time, the use of whole blood is a well-accepted and commonly employed measure without which many modern surgical procedures could not be carried out. -

• Glycolysis • Gluconeogenesis • Glycogen Synthesis
Carbohydrate Metabolism! Wichit Suthammarak – Department of Biochemistry, Faculty of Medicine Siriraj Hospital – Aug 1st and 4th, 2014! • Glycolysis • Gluconeogenesis • Glycogen synthesis • Glycogenolysis • Pentose phosphate pathway • Metabolism of other hexoses Carbohydrate Digestion! Digestive enzymes! Polysaccharides/complex carbohydrates Salivary glands Amylase Pancreas Oligosaccharides/dextrins Dextrinase Membrane-bound Microvilli Brush border Maltose Sucrose Lactose Maltase Sucrase Lactase ‘Disaccharidase’ 2 glucose 1 glucose 1 glucose 1 fructose 1 galactose Lactose Intolerance! Cause & Pathophysiology! Normal lactose digestion Lactose intolerance Lactose Lactose Lactose Glucose Small Intestine Lactase lactase X Galactose Bacteria 1 glucose Large Fermentation 1 galactose Intestine gases, organic acid, Normal stools osmotically Lactase deficiency! active molecules • Primary lactase deficiency: อาการ! genetic defect, การสราง lactase ลด ลงเมออายมากขน, พบมากทสด! ปวดทอง, ถายเหลว, คลนไสอาเจยนภาย • Secondary lactase deficiency: หลงจากรบประทานอาหารทม lactose acquired/transient เชน small bowel เปนปรมาณมาก เชนนม! injury, gastroenteritis, inflammatory bowel disease! Absorption of Hexoses! Site: duodenum! Intestinal lumen Enterocytes Membrane Transporter! Blood SGLT1: sodium-glucose transporter Na+" Na+" •! Presents at the apical membrane ! of enterocytes! SGLT1 Glucose" Glucose" •! Co-transports Na+ and glucose/! Galactose" Galactose" galactose! GLUT2 Fructose" Fructose" GLUT5 GLUT5 •! Transports fructose from the ! intestinal lumen into enterocytes! -

The Role of NF-Κb and C/EBP Factors During Pathogen-Mediated
The role of NF-B and C/EBP factors during pathogen-mediated activation of bovine interleukin 8 and beta-defensin in mammary epithelial cells Inaugural dissertation for the academic degree Doctor rerum naturalium of Mathematisch-Naturwissenschaftlichen Fakultät Universität Rostock By Shuzhen Liu (M. Sc.), born on March-02-1972, in Shanxi Province, China From the Forschungsinstitut für die Biologie landwirtschaftlicher Nutztiere in Dummerstorf Rostock 2009 URN: urn:nbn:de:gbv:28-diss2009-0185-5 Dean: Prof. Dr. Hendrik Schubert Reviewers: 1. Prof. Dr. Hans-Martin Seyfert Research Unit molecular biology, Research Institute for the Biology of Farm Animals, Wilhelm-Stahl-Allee 2, D-18196 Dummerstorf, Germany 2. Prof. Dr. Dieter G. Weiss Division of animal Physiology, Institute of Cell Biology and Biosystems Technology, University of Rostock, Albert-Einstein-Strasse 3, 18059 Rostock, Germany 3. PD Dr. Ulrike Gimsa Research Unit Behavioural Physiology, Research Institute for the Biology of Farm Animals, Wilhelm-Stahl-Allee 2, D-18196 Dummerstorf, Germany Date of defense: October 19th, 2009 Table of Contents TABLE OF CONTENTS 1. INTRODUCTION..................................................................................................................1 1.1 Mastitis as a challenge in general immunology..................................................................1 1.2 Innate immunity of the bovine mammary gland ................................................................2 1.3 Toll-like receptors (TLRs): main receptors perceiving the pathogen -

Biochemistry and the Genomic Revolution 1.1
Dedication About the authors Preface Tools and Techniques Clinical Applications Molecular Evolution Supplements Supporting Biochemistry, Fifth Edition Acknowledgments I. The Molecular Design of Life 1. Prelude: Biochemistry and the Genomic Revolution 1.1. DNA Illustrates the Relation between Form and Function 1.2. Biochemical Unity Underlies Biological Diversity 1.3. Chemical Bonds in Biochemistry 1.4. Biochemistry and Human Biology Appendix: Depicting Molecular Structures 2. Biochemical Evolution 2.1. Key Organic Molecules Are Used by Living Systems 2.2. Evolution Requires Reproduction, Variation, and Selective Pressure 2.3. Energy Transformations Are Necessary to Sustain Living Systems 2.4. Cells Can Respond to Changes in Their Environments Summary Problems Selected Readings 3. Protein Structure and Function 3.1. Proteins Are Built from a Repertoire of 20 Amino Acids 3.2. Primary Structure: Amino Acids Are Linked by Peptide Bonds to Form Polypeptide Chains 3.3. Secondary Structure: Polypeptide Chains Can Fold Into Regular Structures Such as the Alpha Helix, the Beta Sheet, and Turns and Loops 3.4. Tertiary Structure: Water-Soluble Proteins Fold Into Compact Structures with Nonpolar Cores 3.5. Quaternary Structure: Polypeptide Chains Can Assemble Into Multisubunit Structures 3.6. The Amino Acid Sequence of a Protein Determines Its Three-Dimensional Structure Summary Appendix: Acid-Base Concepts Problems Selected Readings 4. Exploring Proteins 4.1. The Purification of Proteins Is an Essential First Step in Understanding Their Function 4.2. Amino Acid Sequences Can Be Determined by Automated Edman Degradation 4.3. Immunology Provides Important Techniques with Which to Investigate Proteins 4.4. Peptides Can Be Synthesized by Automated Solid-Phase Methods 4.5. -

TITAN IV Immunoelectrophoresis Procedure
TITAN IV Immunoelectrophoresis Procedure Cat. No. 9050 and 9061 Helena Laboratories Titan IV Immunoelectrophoresis (IEP) is intended for barbital-sodium barbital buffer with 0.1% sodium azide added semiquantitative determinations by immunoelectrophoresis. as a preservative. WARNING: FOR IN-VITRO DIAGNOSTIC USE ONLY. This SUMMARY product contains sodium azide. Refer to the sodium azide Immunoelectrophoresis (IEP) combines two techniques, warning. electrophoresis and immunodiffusion. In this two-part procedure, Preparation for Use: The plates are ready for use as proteins in a serum or urine sample are first separated packaged. according to charge by electrophoresis. Then, antisera Storage and Stability: Plates should be stored flat, in the complimentary to the proteins under study are applied to the protective packaging, at 2°C to 8°C and are stable until the plate and allowed to diffuse. When a favorable antigen to expiration date indicated on the label. DO NOT FREEZE antibody ratio exists, a precipitin arc will form on the plate. PLATES OR EXPOSE THEM TO EXCESSIVE HEAT. IEP is used for the diagnosis and differential diagnosis of Signs of Deterioration: The plates should have a smooth, monoclonal gammopathies when using serum or urine clear agarose surface. Discard the plates if they appear 1-4 specimens. The method is also used for a number of other cloudy, show bacterial growth, or if they have been exposed to purposes, including screening for circulating immune freezing (a cracked or bubbled surface) or excessive heat (a complexes, characterization of cryoglobulinemia and dried, thin surface). pyroglobulinemia, recognition and characterization of antibody 2. Antisera for Assay of Immunoglobulins syndromes, and recognition and characterization of the various Antiserum to Human IgG, Cat. -

Download the Abstract Book
1 Exploring the male-induced female reproduction of Schistosoma mansoni in a novel medium Jipeng Wang1, Rui Chen1, James Collins1 1) UT Southwestern Medical Center. Schistosomiasis is a neglected tropical disease caused by schistosome parasites that infect over 200 million people. The prodigious egg output of these parasites is the sole driver of pathology due to infection. Female schistosomes rely on continuous pairing with male worms to fuel the maturation of their reproductive organs, yet our understanding of their sexual reproduction is limited because egg production is not sustained for more than a few days in vitro. Here, we explore the process of male-stimulated female maturation in our newly developed ABC169 medium and demonstrate that physical contact with a male worm, and not insemination, is sufficient to induce female development and the production of viable parthenogenetic haploid embryos. By performing an RNAi screen for genes whose expression was enriched in the female reproductive organs, we identify a single nuclear hormone receptor that is required for differentiation and maturation of germ line stem cells in female gonad. Furthermore, we screen genes in non-reproductive tissues that maybe involved in mediating cell signaling during the male-female interplay and identify a transcription factor gli1 whose knockdown prevents male worms from inducing the female sexual maturation while having no effect on male:female pairing. Using RNA-seq, we characterize the gene expression changes of male worms after gli1 knockdown as well as the female transcriptomic changes after pairing with gli1-knockdown males. We are currently exploring the downstream genes of this transcription factor that may mediate the male stimulus associated with pairing. -
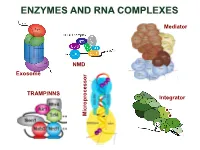
Enzymes and Rna Complexes
ENZYMES AND RNA COMPLEXES Mediator NMD Exosome NMD TRAMP/NNS Integrator Microprocessor RNA PROCESSING and DECAY machinery: RNases Protein Function Characteristics Exonucleases 5’ 3’ Xrn1 cytoplasmic, mRNA degradation processsive Rat1 nuclear, pre-rRNA, sn/snoRNA, pre-mRNA processing and degradation Rrp17/hNol12 nuclear, pre-rRNA processing Exosome 3’ 5’ multisubunit exo/endo complex subunits organized as in bacterial PNPase Rrp44/Dis3 catalytic subunit Exo/PIN domains, processsive Rrp4, Rrp40 pre-rRNA, sn/snoRNA processing, mRNA degradation Rrp41-43, 45-46 participates in NMD, ARE-dependent, non-stop decay Mtr3, Ski4 Mtr4 nuclear helicase cofactor DEAD box Rrp6 (Rrp47) nuclear exonuclease ( Rrp6 BP, cofactor) RNAse D homolog, processsive Ski2,3,7,8 cytoplasmic exosome cofactors. SKI complex helicase, GTPase Other 3’ 5’ Rex1-4 3’-5’ exonucleases, rRNA, snoRNA, tRNA processing RNase D homolog DXO 3’-5’ exonuclease in addition to decapping mtEXO 3’ 5’ mitochondrial degradosome RNA degradation in yeast Suv3/ Dss1 helicase/ 3’-5’ exonuclease DExH box/ RNase II homolog Deadenylation Ccr4/NOT/Pop2 major deadenylase complex (Ccr, Caf, Pop, Not proteins) Ccr4- Mg2+ dependent endonuclease Pan2p/Pan3 additional deadenylases (poliA tail length) RNase D homolog, poly(A) specific nuclease PARN mammalian deadenylase RNase D homolog, poly(A) specific nuclease Endonucleases RNase III -Rnt1 pre-rRNA, sn/snoRNA processing, mRNA degradation dsRNA specific -Dicer, Drosha siRNA/miRNA biogenesis, functions in RNAi PAZ, RNA BD, RNase III domains Ago2 Slicer -

Structure, Function, and Regulation of Intestinal Lactase-Phlorizin Hydrolase and Sucrase- Isomaltase in Health and Disease
Gastrointestinal Functions, edited by Edgard E. Delvin and Michael J. Lentze. Nestle Nutrition Workshop Series, Pediatric Program, Vol. 46. Nestec Ltd.. Vevey/Lippincott Williams & Wilkins. Philadelphia © 2001. Structure, Function, and Regulation of Intestinal Lactase-Phlorizin Hydrolase and Sucrase- Isomaltase in Health and Disease Hassan Y. Nairn Department of Physiological Chemistry, School of Veterinary Medicine Hannover, Hannover, Germany Carbohydrates are essential constituents of the mammalian diet. They occur as oligosaccharides, such as starch and cellulose (plant origin), as glycogen (animal origin), and as free disaccharides such as sucrose (plant) and lactose (animal). These carbohydrates are hydrolyzed in the intestinal lumen by specific enzymes to mono- saccharides before transport across the brush border membrane of epithelial cells into the cell interior. The specificity of the enzymes is determined by the chemical structure of the carbohydrates. The monosaccharide components of most of the known carbohydrates are linked to each other in an a orientation. Examples of this type are starch, glycogen, sucrose, and maltose. (3-Glycosidic linkages are minor. Nevertheless, this type of covalent bond is present in one of the major and most essential carbohydrates in mammalian milk, lactose, which constitutes the primary diet source for the newborn. The enzymes implicated in the digestion of carbohydrates in the intestinal lumen are membrane-bound glycoproteins that are expressed at the apical or microvillus membrane of the enterocytes (1-3). In this chapter, the structural and biosynthetic features of the most important disaccharidases, sucrase-isomaltase and lac- tase-phlorizin hydrolase, and the molecular basis of sugar malabsorption (i.e., en- zyme deficiencies) are discussed. -

CLONING, EXPRESSION ANALYSIS, and TRANSFORMATION VECTOR CONSTRUCTION of DAM HOMOLOGS in PEACH and POPLAR Yuhui Xie Clemson University, [email protected]
Clemson University TigerPrints All Theses Theses 1-2011 CLONING, EXPRESSION ANALYSIS, AND TRANSFORMATION VECTOR CONSTRUCTION OF DAM HOMOLOGS IN PEACH AND POPLAR Yuhui Xie Clemson University, [email protected] Follow this and additional works at: https://tigerprints.clemson.edu/all_theses Part of the Plant Biology Commons Recommended Citation Xie, Yuhui, "CLONING, EXPRESSION ANALYSIS, AND TRANSFORMATION VECTOR CONSTRUCTION OF DAM HOMOLOGS IN PEACH AND POPLAR" (2011). All Theses. 1146. https://tigerprints.clemson.edu/all_theses/1146 This Thesis is brought to you for free and open access by the Theses at TigerPrints. It has been accepted for inclusion in All Theses by an authorized administrator of TigerPrints. For more information, please contact [email protected]. CLONING, EXPRESSION ANALYSIS, AND TRANSFORMATION VECTOR CONSTRUCTION OF DAM HOMOLOGS IN PEACH AND POPLAR _______________________________________________ A Thesis Presented to the Graduate School of Clemson University ________________________________________________ In Partial Fulfillment of the Requirement for the Degree Master of Science Plant and Environmental Sciences _________________________________________________ by Yuhui Xie August 2011 ________________________________________________ Accepted by: Dr. Douglas G. Bielenberg, Committee Chair Dr. Haiying Liang Dr. Hong Luo ABSTRACT Genetic fine mapping and sequencing of the EVG locus in peach [Prunus persica (L.) Batsch] identified six tandem arrayed Dormancy-Associated MADS-box (DAM) genes as candidates for regulating growth cessation and terminal bud formation in the non-dormant evergrowing (evg) mutant. Since the mutant is lacking expression of six genes in the mapped locus, further functional analysis is needed to narrow the list of gene candidates for the non-dormant evg phenotype. Here I report three sets of experiments designed to functionally test DAM genes in peach and their homologs in a model tree, hybrid poplar. -

Clinical Molecular Genetics in the Uk C.1975–C.2000
CLINICAL MOLECULAR GENETICS IN THE UK c.1975–c.2000 The transcript of a Witness Seminar held by the History of Modern Biomedicine Research Group, Queen Mary, University of London, on 5 February 2013 Edited by E M Jones and E M Tansey Volume 48 2014 ©The Trustee of the Wellcome Trust, London, 2014 First published by Queen Mary, University of London, 2014 The History of Modern Biomedicine Research Group is funded by the Wellcome Trust, which is a registered charity, no. 210183. ISBN 978 0 90223 888 6 All volumes are freely available online at www.history.qmul.ac.uk/research/modbiomed/ wellcome_witnesses/ Please cite as: Jones E M, Tansey E M. (eds) (2014) Clinical Molecular Genetics in the UK c.1975–c.2000. Wellcome Witnesses to Contemporary Medicine, vol. 48. London: Queen Mary, University of London. CONTENTS What is a Witness Seminar? v Acknowledgements E M Tansey and E M Jones vii Illustrations and credits ix Abbreviations xi Ancillary guides xiii Introduction Professor Bob Williamson xv Transcript Edited by E M Jones and E M Tansey 1 Appendix 1 Photograph, with key, of delegates attending The Molecular Biology of Thalassaemia conference in Kolimbari, Crete, 1978 88 Appendix 2 Extracts from the University of Leiden postgraduate course Restriction Fragment Length Polymorphisms and Human Genetics, 1982 91 Appendix 3 Archival material of the Clinical Molecular Genetics Society 95 Biographical notes 101 References 113 Index 131 Witness Seminars: Meetings and Publications 143 WHAT IS A WITNESS SEMINAR? The Witness Seminar is a specialized form of oral history, where several individuals associated with a particular set of circumstances or events are invited to meet together to discuss, debate, and agree or disagree about their memories. -

Journal of Biomedical Discovery and Collaboration
View metadata, citation and similar papers at core.ac.uk brought to you by CORE Journal of Biomedical Discovery and provided by PubMed Central Collaboration BioMed Central Case Study Open Access The emergence and diffusion of DNA microarray technology Tim Lenoir*† and Eric Giannella† Address: Jenkins Collaboratory for New Technologies in Society, Duke University, John Hope Franklin Center, 2204 Erwin Road, Durham, North Carolina 27708-0402, USA Email: Tim Lenoir* - [email protected]; Eric Giannella - [email protected] * Corresponding author †Equal contributors Published: 22 August 2006 Received: 09 August 2006 Accepted: 22 August 2006 Journal of Biomedical Discovery and Collaboration 2006, 1:11 doi:10.1186/1747-5333-1-11 This article is available from: http://www.j-biomed-discovery.com/content/1/1/11 © 2006 Lenoir and Giannella; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract The network model of innovation widely adopted among researchers in the economics of science and technology posits relatively porous boundaries between firms and academic research programs and a bi-directional flow of inventions, personnel, and tacit knowledge between sites of university and industry innovation. Moreover, the model suggests that these bi-directional flows should be considered as mutual stimulation of research and invention in both industry and academe, operating as a positive feedback loop. One side of this bi-directional flow – namely; the flow of inventions into industry through the licensing of university-based technologies – has been well studied; but the reverse phenomenon of the stimulation of university research through the absorption of new directions emanating from industry has yet to be investigated in much detail. -

Enzymes in Cloning Part I
˹̀/˺̀/˺̊˼̊ ENZYMES IN CLONING PART I Dr.Sarookhani ˺ ˹̀/˺̀/˺̊˼̊ Cloning --aa definition •• From the Greek --klon,klon, a twig •• An aggregate of the asexually produced progeny of an individual;a group of replicas of all or part of a macromolecule (such as DNA or an antibody) •• An individual grown from a single somatic cell of its parent & genetically identical to it •• Clone: a collection of molecules or cells, all identical to an original molecule or cell Dr.Sarookhani ˻ ˹̀/˺̀/˺̊˼̊ Different types of Cloning 1. Reproductive Cloning 2. Therapeutic Cloning 3. Recombinant DNA Technology or DNA Cloning Dr.Sarookhani ˼ ˹̀/˺̀/˺̊˼̊ DNA CLONING A method for identifying and purifying a particular DNA fragment (clone) of interest from a complex mixture of DNA fragments, and then producing large numbers of the fragment (clone) of interest. Dr.Sarookhani ̊ ˹̀/˺̀/˺̊˼̊ What is genetic engineering • Genetic engineering, also known as recombinant DNA technology, means altering the genes in a living organism to produce a Genetically Modified Organism (GMO) with a new genotype. • Various kinds of genetic modification are possible: inserting a foreign gene from one species into another, forming a transgenic organism; altering an existing gene so that its product is changed; or changing gene expression so that it is translated more often or not at all. Dr.Sarookhani ̋ ˹̀/˺̀/˺̊˼̊ Dr.Sarookhani ̌ ˹̀/˺̀/˺̊˼̊ Dr.Sarookhani ̀ ˹̀/˺̀/˺̊˼̊ Genomic Library Dr.Sarookhani ́ ˹̀/˺̀/˺̊˼̊ Dr.Sarookhani ̂ ˹̀/˺̀/˺̊˼̊ Basic steps in genetic engineering 1.