Original Article the Human Oral Microbiome Database: a Web Accessible Resource for Investigating Oral Microbe Taxonomic and Genomic Information
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Contribution of the Human Microbiome and Proteus Mirabilis to Onset and Progression of Rheumatoid Arthritis: Potential for Targeted Therapy
Internet Journal of Allied Health Sciences and Practice Volume 19 Number 3 Article 6 2021 Contribution of the Human Microbiome and Proteus mirabilis to Onset and Progression of Rheumatoid Arthritis: Potential for Targeted Therapy Jessica Kerpez Nova Southeastern University, [email protected] Marc Kesselman Nova Southeastern University, [email protected] Michelle Demory Beckler Nova Southeastern University, [email protected] Follow this and additional works at: https://nsuworks.nova.edu/ijahsp Part of the Genetic Phenomena Commons, Genetic Processes Commons, Immune System Diseases Commons, Medical Biochemistry Commons, and the Musculoskeletal Diseases Commons Recommended Citation Kerpez J, Kesselman M, Demory Beckler M. Contribution of the Human Microbiome and Proteus mirabilis to Onset and Progression of Rheumatoid Arthritis: Potential for Targeted Therapy. The Internet Journal of Allied Health Sciences and Practice. 2021 Jan 01;19(3), Article 6. This Review Article is brought to you for free and open access by the College of Health Care Sciences at NSUWorks. It has been accepted for inclusion in Internet Journal of Allied Health Sciences and Practice by an authorized editor of NSUWorks. For more information, please contact [email protected]. Contribution of the Human Microbiome and Proteus mirabilis to Onset and Progression of Rheumatoid Arthritis: Potential for Targeted Therapy Abstract The human microbiome has been shown to play a role in the regulation of human health, behavior, and disease. Data suggests that microorganisms that co-evolved within humans have an enhanced ability to prevent the development of a large spectrum of immune-related disorders but may also lead to the onset of conditions when homeostasis is disrupted. -

Health Impact and Therapeutic Manipulation of the Gut Microbiome
Review Health Impact and Therapeutic Manipulation of the Gut Microbiome Eric Banan-Mwine Daliri 1 , Fred Kwame Ofosu 1 , Ramachandran Chelliah 1, Byong Hoon Lee 2,3,* and Deog-Hwan Oh 1 1 Department of Food Science and Biotechnology, Kangwon National University, Chuncheon 200-701, Korea; [email protected] (E.B.-M.D.); [email protected] (F.K.O.); [email protected] (R.C.); [email protected] (D.-H.O.) 2 Department of Microbiology/Immunology, McGill University, Montreal, QC H3A 2B4, Canada 3 SportBiomics, Sacramento Inc., Sacramento, CA 95660, USA * Correspondence: [email protected] Received: 24 March 2020; Accepted: 19 July 2020; Published: 29 July 2020 Abstract: Recent advances in microbiome studies have revealed much information about how the gut virome, mycobiome, and gut bacteria influence health and disease. Over the years, many studies have reported associations between the gut microflora under different pathological conditions. However, information about the role of gut metabolites and the mechanisms by which the gut microbiota affect health and disease does not provide enough evidence. Recent advances in next-generation sequencing and metabolomics coupled with large, randomized clinical trials are helping scientists to understand whether gut dysbiosis precedes pathology or gut dysbiosis is secondary to pathology. In this review, we discuss our current knowledge on the impact of gut bacteria, virome, and mycobiome interactions with the host and how they could be manipulated to promote health. Keywords: microbiome; biomarkers; personalized nutrition; microbes; metagenomics 1. Introduction The human body consists of mammalian cells and many microbial cells (bacteria, viruses, and fungi) which co-exist symbiotically. -

The Human Microbiome, Diet, and Health: Workshop Summary
This PDF is available from The National Academies Press at http://www.nap.edu/catalog.php?record_id=13522 The Human Microbiome, Diet, and Health: Workshop Summary ISBN Leslie Pray, Laura Pillsbury, and Emily Tomayko, Rapporteurs; Food 978-0-309-26585-0 Forum; Food and Nutrition Board; Institute of Medicine 181 pages 6 x 9 PAPERBACK (2013) Visit the National Academies Press online and register for... Instant access to free PDF downloads of titles from the NATIONAL ACADEMY OF SCIENCES NATIONAL ACADEMY OF ENGINEERING INSTITUTE OF MEDICINE NATIONAL RESEARCH COUNCIL 10% off print titles Custom notification of new releases in your field of interest Special offers and discounts Distribution, posting, or copying of this PDF is strictly prohibited without written permission of the National Academies Press. Unless otherwise indicated, all materials in this PDF are copyrighted by the National Academy of Sciences. Request reprint permission for this book Copyright © National Academy of Sciences. All rights reserved. The Human Microbiome, Diet, and Health: Workshop Summary Workshop Summary The Human Microbiome, Diet, and Health Leslie Pray, Laura Pillsbury, and Emily Tomayko, Rapporteurs Food Forum Food and Nutrition Board Copyright © National Academy of Sciences. All rights reserved. The Human Microbiome, Diet, and Health: Workshop Summary THE NATIONAL ACADEMIES PRESS 500 Fifth Street, NW Washington, DC 20001 NOTICE: The project that is the subject of this report was approved by the Govern- ing Board of the National Research Council, whose members are drawn from the councils of the National Academy of Sciences, the National Academy of Engineer- ing, and the Institute of Medicine. This study was supported by Contract Nos. -

Skin Microbiome Analysis for Forensic Human Identification: What Do We Know So Far?
microorganisms Review Skin Microbiome Analysis for Forensic Human Identification: What Do We Know So Far? Pamela Tozzo 1,*, Gabriella D’Angiolella 2 , Paola Brun 3, Ignazio Castagliuolo 3, Sarah Gino 4 and Luciana Caenazzo 1 1 Department of Molecular Medicine, Laboratory of Forensic Genetics, University of Padova, 35121 Padova, Italy; [email protected] 2 Department of Cardiac, Thoracic, Vascular Sciences and Public Health, University of Padova, 35121 Padova, Italy; [email protected] 3 Department of Molecular Medicine, Section of Microbiology, University of Padova, 35121 Padova, Italy; [email protected] (P.B.); [email protected] (I.C.) 4 Department of Health Sciences, University of Piemonte Orientale, 28100 Novara, Italy; [email protected] * Correspondence: [email protected]; Tel.: +39-0498272234 Received: 11 May 2020; Accepted: 8 June 2020; Published: 9 June 2020 Abstract: Microbiome research is a highly transdisciplinary field with a wide range of applications and methods for studying it, involving different computational approaches and models. The fact that different people host radically different microbiota highlights forensic perspectives in understanding what leads to this variation and what regulates it, in order to effectively use microbes as forensic evidence. This narrative review provides an overview of some of the main scientific works so far produced, focusing on the potentiality of using skin microbiome profiling for human identification in forensics. This review was performed following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. The examined literature clearly ascertains that skin microbial communities, although personalized, vary systematically across body sites and time, with intrapersonal differences over time smaller than interpersonal ones, showing such a high degree of spatial and temporal variability that the degree and nature of this variability can constitute in itself an important parameter useful in distinguishing individuals from one another. -

Potential Role of Microbiome in Oncogenesis, Outcome Prediction
Oral Oncology 99 (2019) 104453 Contents lists available at ScienceDirect Oral Oncology journal homepage: www.elsevier.com/locate/oraloncology Review Potential role of microbiome in oncogenesis, outcome prediction and therapeutic targeting for head and neck cancer T ⁎ Ester Orlandia,b, , Nicola Alessandro Iacovellib, Vincenzo Tombolinic, Tiziana Rancatid, Antonella Polimenie, Loris De Ceccof, Riccardo Valdagnia,d,g, Francesca De Felicec a Department of Radiotherapy 1, Fondazione IRCCS Istituto Nazionale dei Tumori di Milano, Milan, Italy b Department of Radiotherapy 2, Fondazione IRCCS Istituto Nazionale dei Tumori di Milano, Milan, Italy c Department of Radiotherapy, Policlinico Umberto I, “Sapienza” University of Rome, Rome, Italy d Prostate Cancer Program, Fondazione IRCCS Istituto Nazionale dei Tumori di Milano, Milan, Italy e Department of Oral and Maxillo Facial Sciences, Policlinico Umberto I, “Sapienza” University of Rome, Italy f Integrated Biology Platform, Department of Applied Research and Technology Development, Fondazione IRCCS Istituto Nazionale dei Tumori di Milano, Milan, Italy g Department of Oncology and Hemato-Oncology, University of Milan, Milan, Italy ARTICLE INFO ABSTRACT Keywords: In the last decade, human microbiome research is rapidly growing involving several fields of clinical medicine Head and neck cancer and population health. Although the microbiome seems to be linked to all sorts of diseases, cancer has the Biomarkers biggest potential to be investigated. Microbiome Following the publication of the National Institute of Health - Human Microbiome Project (NIH-HMP), the Microbiota link between Head and Neck Cancer (HNC) and microbiome seems to be a fast-moving field in research area. Oncogenesis However, robust evidence-based literature is still quite scarce. Nevertheless the relationship between oral mi- Radiotherapy Immunotherapy crobiome and HNC could have important consequences for prevention and early detection of this type of tumors. -

3. Jedna Z Krvných Skupín Systému AB0; 4. Skr. Bukálny. B – Symbo
B – 1. symbol pre bel; 2. chem. značka prvku →bór; 3. jedna z krvných skupín systému AB0; 4. skr. bukálny. B – symbol pre hustotu magnetického toku. B. – skr. pre →Bacillus. B-ALP – skr. angl. bone alcalic phosphatase kostná alkalická fosfatáza. B-bunky – 1. syn. -bunky Langerhansových ostrovčekov; →pankreas; 2. syn. B-lymfocyty; →lymfocyty. B-komplex – multivitamínový prípravok vitamínov B. B-lymfocyty – [B podľa Fabriciovej burzy, imunol. orgánu vtákov] B bunky, druh lymfocytov, kt. sa zúčastňuje na humorálnej imunite (tvorbe protilátok) a niekt. ďalších imunitných funkciách. Povrchové molekuly sú CD 19,20. Receptorom pre antigén je membránový imunoglobulín. B-reťazec – jeden z polypeptidových reťazcov →inzulínu. B-vírus – 1. vírus →hepatitídy B.; 2. vírus zo skupiny Cercopithecine herpes virus 1. B-vlákna →nerv. b – skr. 1. pre barn; 2. skr. angl. born narodený; 3. skr. báza (genet. označenie dĺţky sekvencie nukleotidov, napr. 50 b = sekvencia 50 nukleotidov). b-vlna – pozit. kmit s vyskou amplitúdou v →elektroretinograme nasledujúci po vlne a, prejav komplexnej aktivity vrstvy bipolárnych buniek. – - – predpona označujúca 1. staršie označenie druhého atómu uhlíka v reťazci, na kt. sa pripája hlavná funkčná skupina, napr. kys. -hydroxymaslová správ. kys. 3-hydroxymaslová; 2. špecifická rotácia opticky aktívnej látky, napr. -D-glukóza; 3. orientácia exocyklického atómu al. skupiny, napr. cholest-5-en-3--ol; 4. plazmatický proteín, kt. migruje v elektroforéze v pruhu - lipoproteín; 5. člen série príbuzných chem. látok, najmä série stereoizomérov, izomérov, polymérov al. alotroických foriem, napr. -karotén; 6. -lúč. B2 – staršie označenie pre CD21. B4 – staršie označenie pre CD 19. B19 virus – ľudský parvovírus z čeľade Parvoviridae, značne rozšírený, vyvolávajúci obvykle inaparentné infekcie. -

974-Form.Pdf
California Association for Medical Laboratory Technology Distance Learning Program ANAEROBIC BACTERIOLOGY FOR THE CLINICAL LABORATORY by James I. Mangels, MA, CLS, MT(ASCP) Consultant Microbiology Consulting Services Santa Rosa, CA Course Number: DL-974 3.0 CE/Contact Hour Level of Difficulty: Intermediate © California Association for Medical Laboratory Technology. Permission to reprint any part of these materials, other than for credit from CAMLT, must be obtained in writing from the CAMLT Executive Office. CAMLT is approved by the California Department of Health Services as a CA CLS Accrediting Agency (#0021) and this course is is approved by ASCLS for the P.A.C.E. ® Program (#519) 1895 Mowry Ave, Suite 112 Fremont, CA 94538-1766 Phone: 510-792-4441 FAX: 510-792-3045 Notification of Distance Learning Deadline All continuing education units required to renew your license must be earned no later than the expiration date printed on your license. If some of your units are made up of Distance Learning courses, please allow yourself enough time to retake the test in the event you do not pass on the first attempt. CAMLT urges you to earn your CE units early!. CAMLT Distance Learning Course # DL-974 1 © California Association for Medical Laboratory Technology Outline A. Introduction B. What are anaerobic bacteria? Concepts of anaerobic bacteriology C. Why do we need to identify anaerobes? D. Normal indigenous anaerobic flora; the incidence of anaerobes at various body sites E. Anaerobic infections; most common anaerobic infections F. Specimen collection and transport; acceptance and rejection criteria G. Processing of clinical specimens 1. Microscopic examination 2. -

Population Genetics in the Human Microbiome
Population Genetics in the Human Microbiome Nandita Garud Department of Ecology and Evolutionary Biology, UCLA [email protected] | http://garud.eeb.ucla.edu @nanditagarud The human microbiome is a complex community Bacteria Fungi Viruses Archaea Who is there? What are they doing? Who is there? What are they doing? How are they evolving? Rapid evolution in a simple community Time Richard Lenski and colleagues Do microbiota respond to their environment by evolutionary or ecological processes? Species fluctuations Do microbiota respond to their environment by evolutionary or ecological processes? X Invasion X X X X X X X X X Do microbiota respond to their environment by evolutionary or ecological processes? Evolution Do microbiota respond to their environment by evolutionary or ecological processes? Evolution = change in allele frequency ≠ change in species composition Abundant opportunity for evolution in the human microbiome ~1:1 ratio of human to microbial cells! Sender et al. 2016 PLoS Bio. ~1 billion mutations entering our microbiomes daily! Zhao, Lieberman et al. 2019 Cell Host Microbe Evolution What determines the fate of a new mutation? • Data • Drift and migration • Adaptation • Recombination Population genetic processes can give rise to a range of traits in the microbiome Digestion of food Antibiotic resistance Drug metabolism Hehemann et al. 2010 Nature (e.g. Karami et al. 2007 J. Antimicrob. (Haiser et al. 2013 Science) Chemother.) Ecology AND Evolution Ecology Evolution Distribution of allele Distribution of species frequencies -

In 1683 a Dutchman, Antonie Van Leeuwenhoek, Having Developed
How do we benefit from the microorganisms that live within us? In 1683 a Dutchman, Antonie van Leeuwenhoek, having developed microscopes many times more powerful than any before, examined the white matter from his teeth and observed, ‘the number of animals in the scruff of a man’s teeth are so many that I believe they exceed the number of men in a kingdom.’ After his death in 1723, no other scientists were able to see these ‘animalcules’ without the impressive magnification unique to Leeuwenhoek’s microscopes (bequeathed to, and promptly 2,3 lost by, the Royal Society) leaving his discovery to be dismissed for almost 150 years . Microbes only reclaimed the scientific spotlight 1860s when Louis Pasteur proved they could cause disease. However, this was the genesis of ‘germ theory,’ which indiscriminately cast all our resident bacteria as malicious invaders, and ensured that the microorganisms living within our bodies became known only as enemies to our health. Since then, modern medicine has focused on the war against microbes - through sanitation, antiseptics, and the miraculous development of antibiotics. The idea that these supposed “enemies” could actually be good for us only emerged this century, gaining prominence when the Human Microbiome Project in 2007 - a 5 year program to sequence the genomes of microbial populations of 300 healthy Americans - helped to unveil the myriad ways our resident bacteria benefit us. Our body’s flora has frequently been described as an entire organ. It harbours a diverse community of intricately connected microbes in a precarious state of symbiosis. In exchange for a favourable habitat, these microbes fulfil many functions we can’t perform ourselves: helping to metabolise otherwise indigestible compounds, regulating our immune response, and even influencing brain activity. -
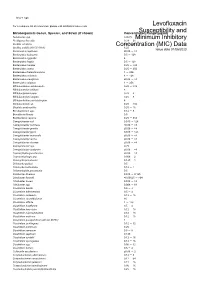
Susceptibility and Resistance Data
toku-e logo For a complete list of references, please visit antibiotics.toku-e.com Levofloxacin Microorganism Genus, Species, and Strain (if shown) Concentration Range (μg/ml)Susceptibility and Aeromonas spp. 0.0625 Minimum Inhibitory Alcaligenes faecalis 0.39 - 25 Bacillus circulans Concentration0.25 - 8 (MIC) Data Bacillus subtilis (ATCC 6051) 6.25 Issue date 01/06/2020 Bacteroides capillosus ≤0.06 - >8 Bacteroides distasonis 0.5 - 128 Bacteroides eggerthii 4 Bacteroides fragilis 0.5 - 128 Bacteroides merdae 0.25 - >32 Bacteroides ovatus 0.25 - 256 Bacteroides thetaiotaomicron 1 - 256 Bacteroides uniformis 4 - 128 Bacteroides ureolyticus ≤0.06 - >8 Bacteroides vulgatus 1 - 256 Bifidobacterium adolescentis 0.25 - >32 Bifidobacterium bifidum 8 Bifidobacterium breve 0.25 - 8 Bifidobacterium longum 0.25 - 8 Bifidobacterium pseudolongum 8 Bifidobacterium sp. 0.25 - >32 Bilophila wadsworthia 0.25 - 16 Brevibacterium spp. 0.12 - 8 Brucella melitensis 0.5 Burkholderia cepacia 0.25 - 512 Campylobacter coli 0.015 - 128 Campylobacter concisus ≤0.06 - >8 Campylobacter gracilis ≤0.06 - >8 Campylobacter jejuni 0.015 - 128 Campylobacter mucosalis ≤0.06 - >8 Campylobacter rectus ≤0.06 - >8 Campylobacter showae ≤0.06 - >8 Campylobacter spp. 0.25 Campylobacter sputorum ≤0.06 - >8 Capnocytophaga ochracea ≤0.06 - >8 Capnocytophaga spp. 0.006 - 2 Chlamydia pneumonia 0.125 - 1 Chlamydia psittaci 0.5 Chlamydia trachomatis 0.12 - 1 Chlamydophila pneumonia 0.5 Citrobacter diversus 0.015 - 0.125 Citrobacter freundii ≤0.00625 - >64 Citrobacter koseri 0.015 - -

(12) United States Patent (10) Patent No.: US 8,501,463 B2 Cox Et Al
USOO85O1463B2 (12) United States Patent (10) Patent No.: US 8,501,463 B2 Cox et al. (45) Date of Patent: Aug. 6, 2013 (54) ANAEROBC PRODUCTION OF HYDROGEN (56) References Cited AND OTHER CHEMICAL PRODUCTS U.S. PATENT DOCUMENTS (75) Inventors: Marion E. Cox, Morgan Hill, CA (US); 5,350,685 A 9/1994 Taguchi et al. Laura M. Nondorf, Morgan Hill, CA 5,464,539 A 11/1995 Ueno et al. 6,090,266 A 7/2000 Roychowdhury (US); Steven M. Cox, Morgan Hill, CA 6,251,643 B1 6/2001 Hansen et al. (US) 6,299,774 B1 * 10/2001 Ainsworth et al. ........... 210,603 6,342,378 B1 1/2002 Zhang et al. (73) Assignee: Anaerobe Systems, Morgan Hill, CA 6,569,332 B2 * 5/2003 Ainsworth et al. ........... 210,603 2004/0050778 A1 3/2004 Noike et al. (US) 2004/O115782 A1 6/2004 Paterek (*) Notice: Subject to any disclaimer, the term of this FOREIGN PATENT DOCUMENTS patent is extended or adjusted under 35 WO WO-2006-119052 A2 11/2006 U.S.C. 154(b) by 1347 days. OTHER PUBLICATIONS (21) Appl. No.: 11/912,881 Liu et al., 2004. Effects of Culture and Medium Conditions on Hydro gen Production from Starch Using Anaerobic Bacteria. Journal of (22) PCT Fled: Apr. 27, 2006 Bioscience and Bioengineering, vol. 98, No. 4, pp. 251-256.* Zhang et al., Distributed Computer Control of Penicillin Fermenta (86) PCT NO.: PCT/US2OO6/O16332 tion Industrial Production. Proceedings of the IEEE International Conference on Industrial Technology, 1996, pp. 52-56.* S371 (c)(1), New Brunswick, an eppenforf Company, pp. -

Kin Selection Explains the Evolution of Cooperation in the Gut Microbiota
Kin selection explains the evolution of cooperation in the gut microbiota Camille Simoneta,1 and Luke McNallya,b aInstitute of Evolutionary Biology, School of Biological Sciences, University of Edinburgh, Edinburgh EH9 3FL, United Kingdom; and bCentre for Synthetic and Systems Biology, School of Biological Sciences, University of Edinburgh, Edinburgh EH9 3BF, United Kingdom Edited by Joan E. Strassmann, Washington University in St. Louis, St. Louis, MO, and approved December 17, 2020 (received for review July 29, 2020) Through the secretion of “public goods” molecules, microbes coop- relatedness, because it limits those indirect fitness benefits (21) eratively exploit their habitat. This is known as a major driver of (Fig. 1A). Hamilton’s theory generates a prediction of great gen- the functioning of microbial communities, including in human dis- erality: All else being equal, increased relatedness should lead ease. Understanding why microbial species cooperate is therefore to more cooperation. Contrary to predictions based on specific crucial to achieve successful microbial community management, mechanisms [e.g., pleiotropy (22) or greenbeard genes (23, 24)] such as microbiome manipulation. A leading explanation is that of or that apply to a limited amount of taxa [e.g., particular sce- Hamilton’s inclusive-fitness framework. A cooperator can indirectly narios calling upon preadaptations (25, 26)], the generality of transmit its genes by helping the reproduction of an individual car- Hamilton’s prediction is useful in that it identifies a unifying rying similar genes. Therefore, all else being equal, as relatedness parameter (27). In the context of mastering MCs that are hugely among individuals increases, so should cooperation.