Stochastic Turing Patterns in a Synthetic Bacterial Population
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Title: Fractals Topics: Symmetry, Patterns in Nature, Biomimicry Related Disciplines: Mathematics, Biology Objectives: A. Learn
Title: Fractals Topics: symmetry, patterns in nature, biomimicry Related Disciplines: mathematics, biology Objectives: A. Learn about how fractals are made. B. Think about the mathematical processes that play out in nature. C. Create a hands-on art project. Lesson: A. Introduction (20 minutes) Fractals are sets in mathematics based on repeated processes at different scales. Why do we care? Fractals are a part of nature, geometry, algebra, and science and are beautiful in their complexity. Fractals come in many types: branching, spiral, algebraic, and geometric. Branching and spiral fractals can be found in nature in many different forms. Branching is seen in trees, lungs, snowflakes, lightning, and rivers. Spirals are visible in natural forms such as hurricanes, shells, liquid motion, galaxies, and most easily visible in plants like flowers, cacti, and Romanesco Figure 1: Romanesco (Figure 1). Branching fractals emerge in a linear fashion. Spirals begin at a point and expand out in a circular motion. A cool demonstration of branching fractals can be found at: http://fractalfoundation.org/resources/what-are-fractals/. Algebraic fractals are based on surprisingly simple equations. Most famous of these equations is the Mandelbrot Set (Figure 2), a plot made from the equation: The most famous of the geometric fractals is the Sierpinski Triangle: As seen in the diagram above, this complex fractal is created by starting with a single triangle, then forming another inside one quarter the size, then three more, each one quarter the size, then 9, 27, 81, all just one quarter the size of the triangle drawn in the previous step. -

Branching in Nature Jennifer Welborn Amherst Regional Middle School, [email protected]
University of Massachusetts Amherst ScholarWorks@UMass Amherst Patterns Around Us STEM Education Institute 2017 Branching in Nature Jennifer Welborn Amherst Regional Middle School, [email protected] Wayne Kermenski Hawlemont Regional School, [email protected] Follow this and additional works at: https://scholarworks.umass.edu/stem_patterns Part of the Biology Commons, Physics Commons, Science and Mathematics Education Commons, and the Teacher Education and Professional Development Commons Welborn, Jennifer and Kermenski, Wayne, "Branching in Nature" (2017). Patterns Around Us. 2. Retrieved from https://scholarworks.umass.edu/stem_patterns/2 This Article is brought to you for free and open access by the STEM Education Institute at ScholarWorks@UMass Amherst. It has been accepted for inclusion in Patterns Around Us by an authorized administrator of ScholarWorks@UMass Amherst. For more information, please contact [email protected]. Patterns Around Us: Branching in Nature Teacher Resource Page Part A: Introduction to Branching Massachusetts Frameworks Alignment—The Nature of Science • Overall, the key criterion of science is that it provide a clear, rational, and succinct account of a pattern in nature. This account must be based on data gathering and analysis and other evidence obtained through direct observations or experiments, reflect inferences that are broadly shared and communicated, and be accompanied by a model that offers a naturalistic explanation expressed in conceptual, mathematical, and/or mechanical terms. Materials: -

Art on a Cellular Level Art and Science Educational Resource
Art on a Cellular Level Art and Science Educational Resource Phoenix Airport Museum Educators and Parents, With foundations in art, geometry and plant biology, the objective of this lesson is to recognize patterns and make connections between the inexhaustible variety of life on our planet. This educational resource is geared for interaction with students of all ages to support the understanding between art and science. It has been designed based on our current exhibition, Art on a Cellular Level, on display at Sky Harbor. The questions and activities below were created to promote observation and curiosity. There are no wrong answers. You may print this PDF to use as a workbook or have your student refer to the material online. We encourage educators to expand on this art and science course to create a lesson plan. If you enjoy these activities and would like to investigate further, check back for new projects each week (three projects total). We hope your student will have fun with this and make an art project to share with us. Please send an image of your student’s artwork to [email protected] or hashtag #SkyHarborArts for an opportunity to be featured on Phoenix Sky Harbor International Airport’s social media. Art on a Cellular Level exhibition Sky Harbor, Terminal 4, level 3 Gallery Art is a lens through which we view the world. It can be a tool for storytelling, expressing cultural values and teaching fundamentals of math, technology and science in a visual way. The Terminal 4 gallery exhibition, Art on a Cellular Level, examines the intersections between art and science. -
Bios and the Creation of Complexity
BIOS AND THE CREATION OF COMPLEXITY Prepared by Hector Sabelli and Lazar Kovacevic Chicago Center for Creative Development http://creativebios.com BIOS, A LINK BETWEEN CHAOS AND COMPLEXITY BIOS is a causal and creative process that follows chaos in sequences of patterns of increasing complexity. Bios was first identified as the pattern of heartbeat intervals, and it has since then been found in a wide variety of processes ranging in size from Schrodinger’s wave function to the temporal distribution of galaxies, and ranging in complexity from physics to economics to music. This tutorial provides concise descriptions of (1) bios, (2) time series analyses software to identify bios in empirical data, (3) biotic patterns in nature, (4) biotic recursions, (5) biotic feedback, (6) Bios Theory of Evolution, and (7) biotic strategies for human action in scientific, clinical, economic and sociopolitical settings. Time series of heartbeat intervals (RRI), and of biotic and chaotic series generated with mathematical recursions of bipolar feedback. Heartbeats are the prototype of bios. Turbulence is the prototype of chaos. 1 1. BIOS BIOS is an expansive process with chaotic features generated by feedback and characterized by features of creativity. Process: Biotic patterns are sequences of actions or states. Expansive: Biotic patterns continually expand in their diversity and often in their range. This is significant, as natural processes expand, in contrast to convergence to equilibrium, periodic, or chaotic attractors. Expanding processes range from the universe to viruses, and include human populations, empires, ideas and cultures. Chaotic: Biotic series are aperiodic and generated causally; mathematically generated bios is extremely sensitive to initial conditions. -

Nanostructures De Titanate De Baryum: Modélisation,Simulations
Nanostructures de titanate de Baryum : modélisation,simulations numériques et étude expérimentale. Gentien Thorner To cite this version: Gentien Thorner. Nanostructures de titanate de Baryum : modélisation,simulations numériques et étude expérimentale.. Autre. Université Paris Saclay (COmUE), 2016. Français. NNT : 2016SACLC093. tel-01427275 HAL Id: tel-01427275 https://tel.archives-ouvertes.fr/tel-01427275 Submitted on 5 Jan 2017 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. NNT : 2016SACLC093 THÈSE DE DOCTORAT DE L’UNIVERSITÉ PARIS-SACLAY PRÉPARÉE À “CENTRALESUPÉLEC” ECOLE DOCTORALE N° (573) Interfaces : approches interdisciplinaires, fondements, applications et innovation Physique Par Monsieur Gentien Thorner BARIUM TITANATE NANOSTRUCTURES: modeling, numerical simulations and experiments Thèse présentée et soutenue à Châtenay-Malabry, le 28 novembre 2016: Composition du Jury : Philippe Papet Professeur, Université de Montpellier Président Laurent Bellaiche Professeur, Université d'Arkansas Rapporteur Gregory Geneste Ingénieur de Recherche, CEA Rapporteur Christine Bogicevic Ingénieur de Recherche, CentraleSupélec Examinatrice Jean-Michel Kiat Directeur de Recherche, CentraleSupélec Directeur de thèse 3 Remerciements À un moment relativement crucial d’une thèse, quelques pensées iraient tout d’abord vers son directeur Jean-Michel Kiat pour un soutien indéfectible pendant une période s’étalant d’un premier contact lors d’un stage de recherche en deux mille dix jusqu’à la soutenance en deux mille seize. -

Pattern Formation in Nature: Physical Constraints and Self-Organising Characteristics
Pattern formation in nature: Physical constraints and self-organising characteristics Philip Ball ________________________________________________________________ Abstract The formation of patterns is apparent in natural systems ranging from clouds to animal markings, and from sand dunes to the intricate shells of microscopic marine organisms. Despite the astonishing range and variety of such structures, many seem to have analogous features: the zebra’s stripes put us in mind of the ripples of blown sand, for example. In this article I review some of the common patterns found in nature and explain how they are typically formed through simple, local interactions between many components of a system – a form of physical computation that gives rise to self- organisation and emergent structures and behaviours. ________________________________________________________________ Introduction When the naturalist Joseph Banks first encountered Fingal’s Cave on the Scottish island of Staffa, he was astonished by the quasi-geometric, prismatic pillars of rock that flank the entrance. As Banks put it, Compared to this what are the cathedrals or palaces built by men! Mere models or playthings, as diminutive as his works will always be when compared with those of nature. What now is the boast of the architect! Regularity, the only part in which he fancied himself to exceed his mistress, Nature, is here found in her possession, and here it has been for ages undescribed. This structure has a counterpart on the coast of Ireland: the Giant’s Causeway in County Antrim, where again one can see the extraordinarily regular and geometric honeycomb structure of the fractured igneous rock (Figure 1). When we make an architectural pattern like this, it is through careful planning and construction, with each individual element cut to shape and laid in place. -

Exploratory Modelling and the Case of Turing Patterns
to appear in: Christian, A., Hommen, D., Retzlaff, N., Schurz, G. (Eds.): Philosophy of Science. Between the Natural Sciences, the Social Sciences, and the Humanities, New York: Springer 2018. Models in Search of Targets: Exploratory Modelling and the Case of Turing Patterns Axel Gelfert Department of Philosophy, Literature, History of Science & Technology Technical University of Berlin Straße des 17. Juni 135, 10623 Berlin email: [email protected] Abstract Traditional frameworks for evaluating scientific models have tended to downplay their exploratory function; instead they emphasize how models are inherently intended for specific phenomena and are to be judged by their ability to predict, reproduce, or explain empirical observations. By contrast, this paper argues that exploration should stand alongside explanation, prediction, and representation as a core function of scientific models. Thus, models often serve as starting points for future inquiry, as proofs of principle, as sources of potential explanations, and as a tool for reassessing the suitability of the target system (and sometimes of whole research agendas). This is illustrated by a case study of the varied career of reaction- diffusion models in the study of biological pattern formation, which was initiated by Alan Turing in a classic 1952 paper. Initially regarded as mathematically elegant, but biologically irrelevant, demonstrations of how, in principle, spontaneous pattern formation could occur in an organism, such Turing models have only recently rebounded, thanks to advances in experimental techniques and computational methods. The long-delayed vindication of Turing’s initial model, it is argued, is best explained by recognizing it as an exploratory tool (rather than as a purported representation of an actual target system). -

Fractal Art: Closer to Heaven? Modern Mathematics, the Art of Nature, and the Nature of Art
Fractal Art: Closer to Heaven? Modern Mathematics, the art of Nature, and the nature of Art Charalampos Saitis Music & Media Technologies Dept. of Electronic & Electrical Engineering University of Dublin, Trinity College Printing House, Trinity College Dublin 2, Ireland [email protected] Abstract Understanding nature has always been a reference point for both art and science. Aesthetics have put nature at the forefront of artistic achievement. Artworks are expected to represent nature, to work like it. Science has likewise been trying to explain the very laws that determine nature. Technology has provided both sides with the appropriate tools towards their common goal. Fractal art stands right at the heart of the art-science-technology triangle. This paper examines the new perspectives brought to art by fractal geometry and chaos theory and how the study of the fractal character of nature offers promising possibilities towards art’s mission. Introduction “When I judge art, I take my painting and put it next to a God-made object like a tree or flower. If it clashes, it is not art.” Paul Cézanne [1] Mathematics has always been affecting art. Mathematical concepts such as the golden ratio, the platonic solids and the projective geometry have been widely used by painters and sculptors whereas the Pythagorean arithmetic perception of harmony dominates western music to this very day. Infinity and probability theory inspired artists such as M. C. Escher and composers such as Iannis Xenakis forming trends in contemporary art and music. The evolution of technology created new areas of intersection between mathematics and art or music: digital art, computer music and new media. -

Lagrangian Coherent Structures
FL47CH07-Haller ARI 24 November 2014 9:57 Lagrangian Coherent Structures George Haller Institute of Mechanical Systems, ETH Zurich,¨ 8092 Zurich,¨ Switzerland; email: [email protected] Annu. Rev. Fluid Mech. 2015. 47:137–62 Keywords First published online as a Review in Advance on mixing, turbulence, transport, invariant manifolds, nonlinear dynamics August 28, 2014 Access provided by 80.218.229.121 on 01/08/15. For personal use only. The Annual Review of Fluid Mechanics is online at Abstract Annu. Rev. Fluid Mech. 2015.47:137-162. Downloaded from www.annualreviews.org fluid.annualreviews.org Typical fluid particle trajectories are sensitive to changes in their initial con- This article’s doi: ditions. This makes the assessment of flow models and observations from 10.1146/annurev-fluid-010313-141322 individual tracer samples unreliable. Behind complex and sensitive tracer Copyright c 2015 by Annual Reviews. patterns, however, there exists a robust skeleton of material surfaces, La- All rights reserved grangian coherent structures (LCSs), shaping those patterns. Free from the uncertainties of single trajectories, LCSs frame, quantify, and even forecast key aspects of material transport. Several diagnostic quantities have been proposed to visualize LCSs. More recent mathematical approaches iden- tify LCSs precisely through their impact on fluid deformation. This review focuses on the latter developments, illustrating their applications to geo- physical fluid dynamics. 137 FL47CH07-Haller ARI 24 November 2014 9:57 1. INTRODUCTION Material coherence emerges ubiquitously in fluid flows around us (Figure 1). It admits distinct signatures in virtually any diagnostic scalar field or reduced-order model associated with these flows. -

Regulation of Phyllotaxis
CORE Metadata, citation and similar papers at core.ac.uk Published in "International Journal of Developmental Biology 49: 539-546, 2005" Provided by RERO DOC Digital Library which should be cited to refer to this work. Regulation of phyllotaxis DIDIER REINHARDT* Plant Biology, Department of Biology, Fribourg, Switzerland ABSTRACT Plant architecture is characterized by a high degree of regularity. Leaves, flowers and floral organs are arranged in regular patterns, a phenomenon referred to as phyllotaxis. Regular phyllotaxis is found in virtually all higher plants, from mosses, over ferns, to gymnosperms and angiosperms. Due to its remarkable precision, its beauty and its accessibility, phyllotaxis has for centuries been the object of admiration and scientific examination. There have been numerous hypotheses to explain the nature of the mechanistic principle behind phyllotaxis, however, not all of them have been amenable to experimental examination. This is due mainly to the delicacy and small size of the shoot apical meristem, where plant organs are formed and the phyllotactic patterns are laid down. Recently, the combination of genetics, molecular tools and micromanipulation has resulted in the identification of auxin as a central player in organ formation and positioning. This paper discusses some aspects of phyllotactic patterns found in nature and summarizes our current understanding of the regulatory mechanism behind phyllotaxis. KEY WORDS: phyllotaxis, meristem, auxin, Arabidopsis, tomato Introduction formed simultaneously. Spiral phyllotaxis is the most common pattern and is found in model plant species such as Arabidopsis, The architecture of plants is highly regular. For example, flowers tobacco and tomato. An important model system for the study of exhibit characteristic symmetric arrangement of their organs; the distichous phyllotaxis is maize. -
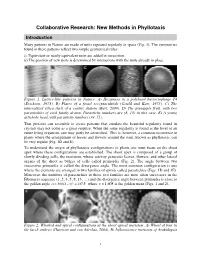
Collaborative Research: New Methods in Phyllotaxis
Collaborative Research: New Methods in Phyllotaxis Introduction Many patterns in Nature are made of units repeated regularly in space (Fig. 1). The symmetries found in these patterns reflect two simple geometrical rules: i) Equivalent or nearly equivalent units are added in succession. ii) The position of new units is determined by interactions with the units already in place. Figure 1. Lattice-like patterns in Nature. A) Hexamers in a polyhead bacteriophage T4 (Erickson, 1973). B) Plates of a fossil receptaculitids (Gould and Katz, 1975). C) The mineralized silica shell of a centric diatom (Bart, 2000). D) The pineapple fruit, with two parastichies of each family drawn. Parastichy numbers are (8, 13) in this case. E) A young artichoke head, with parastichy numbers (34, 55). That proteins can assemble to create patterns that emulate the beautiful regularity found in crystals may not come as a great surprise. When the same regularity is found at the level of an entire living organism, one may justly be astonished. This is, however, a common occurrence in plants where the arrangement of leaves and flowers around the stem, known as phyllotaxis, can be very regular (Fig. 1D and E). To understand the origin of phyllotactic configurations in plants one must focus on the shoot apex where these configurations are established. The shoot apex is composed of a group of slowly dividing cells, the meristem, whose activity generates leaves, flowers, and other lateral organs of the shoot as bulges of cells called primordia (Fig. 2). The angle between two successive primordia is called the divergence angle. -

Vegetative Turing Pattern Formation: a Historical Perspective
Vegetative Turing Pattern Formation: A Historical Perspective Bonni J. Kealy* and David J. Wollkind Washington State University Department of Mathematics Pullman, WA 99164-3113 Joint Mathematics Meetings AMS-ALS Special Session on the Life and Legacy of Alan Turing January 2012 Bonni J. Kealy* and David J. Wollkind Washington State University Vegetative Turing Pattern Formation: A Historical Perspective Alan Turing Born Alan Mathison Turing 23 June 1912 Maida Vale, London, England Died 7 June 1954 (aged 41) Wilmslow, Chesire, England Figure: Turing at the time of his election to Fellowship of the Royal Society Bonni J. Kealy* and David J. Wollkind Washington State University Vegetative Turing Pattern Formation: A Historical Perspective Alan Turing: World Class Distance Runner Alan Turing achieved world-class Marathon standards. His best time of 2 hours, 46 minutes, 3 seconds, was only 11 minutes slower than the winner in the 1948 Olympic Games. In a 1948 cross-country race he finished ahead of Tom Richards who was to win the silver medal in the Olympics. Figure: From The Times, 25 August 1947 Figure: Running in 1946 Bonni J. Kealy* and David J. Wollkind Washington State University Vegetative Turing Pattern Formation: A Historical Perspective Paper: The Chemical Basis of Morphogenesis Figure: An example of a `dappled' Figure: Philosophical Transactions of pattern as resulting from a type (a) the Royal Society of London. Series B, morphogen system. A marker of unit Biological Sciences, Vol. 237, No. 641. length is shown. (Aug. 14, 1952), pp. 37-72. Bonni J. Kealy* and David J. Wollkind Washington State University Vegetative Turing Pattern Formation: A Historical Perspective Examples Bonni J.