Automated Thalamic Nuclei Segmentation Using Multi-Planar Cascaded Convolutional Neural Networks
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Human Thalamus Is an Integrative Hub for Functional Brain Networks
5594 • The Journal of Neuroscience, June 7, 2017 • 37(23):5594–5607 Behavioral/Cognitive The Human Thalamus Is an Integrative Hub for Functional Brain Networks X Kai Hwang, Maxwell A. Bertolero, XWilliam B. Liu, and XMark D’Esposito Helen Wills Neuroscience Institute and Department of Psychology, University of California, Berkeley, Berkeley, California 94720 The thalamus is globally connected with distributed cortical regions, yet the functional significance of this extensive thalamocortical connectivityremainslargelyunknown.Byperforminggraph-theoreticanalysesonthalamocorticalfunctionalconnectivitydatacollected from human participants, we found that most thalamic subdivisions display network properties that are capable of integrating multi- modal information across diverse cortical functional networks. From a meta-analysis of a large dataset of functional brain-imaging experiments, we further found that the thalamus is involved in multiple cognitive functions. Finally, we found that focal thalamic lesions in humans have widespread distal effects, disrupting the modular organization of cortical functional networks. This converging evidence suggests that the human thalamus is a critical hub region that could integrate diverse information being processed throughout the cerebral cortex as well as maintain the modular structure of cortical functional networks. Key words: brain networks; diaschisis; functional connectivity; graph theory; thalamus Significance Statement The thalamus is traditionally viewed as a passive relay station of information from sensory organs or subcortical structures to the cortex. However, the thalamus has extensive connections with the entire cerebral cortex, which can also serve to integrate infor- mation processing between cortical regions. In this study, we demonstrate that multiple thalamic subdivisions display network properties that are capable of integrating information across multiple functional brain networks. Moreover, the thalamus is engaged by tasks requiring multiple cognitive functions. -

The Thalamus
212 Neuroanatomy Reflection Corner The Thalamus Sagar Karia1 1Specialty Medical Officer, Department of Psychiatry, Lokmanya Tilak Municipal Medical College, Mumbai. E-mail – [email protected] INTRODUCTION Word Thalamus from Greek origin meaning “inner room” or “chamber”. Is an egg shaped mass of grey matter forming part of Diencephalon and forming lateral wall of 3rd ventricle. Narrow anterior end is directed medially while broad posterior end is directed laterally. Its long axis is 300 oblique to midline. Size is 3.5cm x 1.5cm. Covering its lateral surface is the external medullary lamina consisting of thalamocortical and corticothalamic fibres. Internal medullary lamina consists mainly of internuclear thalamic connections. It is 'Y' shaped and divides thalamus into 3 different nuclear masses. Thalamic Nuclei Anterior Group: portion between diverse limbs of 'Y'. It contains Anterior nuclei. Medial Group: part of thalamus lying on medial side of stem of 'Y'. It contains intralaminar nuclei, centromedian nuclei, medial nuclei and midline nuclei. Lateral Group: part lying on lateral side of stem of 'Y'. It is divided into 2 groups- ventral group and dorsal group. Ventral group contains ventroanterior, venterolateral, and venteroposterior nuclei and most posteriorly medial and lateral geniculate bodies. Venteroposterior nuclei is divided into venteroposteriorlateral and venteroposteriormedial nuclei. Dorsal group contains pulvinar, lateral posterior and lateral dorsal nuclei. Indian Journal of Mental Health 2015; 2(2) 213 Connections of the Thalamus Functionally divided into extrinsic and intrinsic nuclei. Extrinsic nuclei are cortical relay nuclei and receive afferent fibres from extrathalamic sources. Axons of these cells are distributed to primary cortical areas- pre and post central cortices, visual and auditory cortical areas. -

The Primate Pulvinar Nuclei: Vision and Action
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Repositorio da Universidade da Coruña Trends in Neurosciences, vol. 23, issue 1: 35-39 The primate pulvinar nuclei: vision and action Kenneth L. Grieve, Carlos Acuña and Javier Cudeiro Abstract The pulvinar nuclei of the thalamus are proportionately larger in higher mammals, particularly in primates, and account for a quarter of the total mass. Traditionally, these nuclei have been divided into oral (somatosensory), superior and inferior (both visual) and medial (visual, multi-sensory) divisions. With reciprocal connections to vast areas of cerebral cortex, and input from the colliculus and retina, they occupy an analogous position in the extra- striate visual system to the lateral geniculate nucleus in the primary visual pathway, but deal with higher-order visual and visuomotor transduction. With a renewed recent interest in this thalamic nuclear collection, and growth in our knowledge of the cortex with which it communicates, perhaps the time is right to look to new dimensions in the pulvinar code. Keywords: Primate; Pulvinar; Reference frame; Salience; Cortico-thalamic; Spatial attention The pulvinar nuclei of the thalamus lie posterior, medial and dorsal to their much better known cousin, the lateral geniculate nucleus, and ‘cover’ the underlying superior colliculus (SC). In the same way as the lateral geniculate (‘knee-like’) nucleus curves around the rising optic tract, the pulvinar (‘cushion’) forms a larger and more-diffuse, but recognizable, mass around the axonal tract that arises from the SC, the brachium of the SC (see Fig. 1). The original four nuclei of the macaque pulvinar were defined in early studies on purely anatomical grounds1 and 2. -

Magnetic Resonance Imaging of Mediodorsal, Pulvinar, and Centromedian Nuclei of the Thalamus in Patients with Schizophrenia
ORIGINAL ARTICLE Magnetic Resonance Imaging of Mediodorsal, Pulvinar, and Centromedian Nuclei of the Thalamus in Patients With Schizophrenia Eileen M. Kemether, MD; Monte S. Buchsbaum, MD; William Byne, MD, PhD; Erin A. Hazlett, PhD; Mehmet Haznedar, MD; Adam M. Brickman, MPhil; Jimcy Platholi, MA; Rachel Bloom Background: Postmortem and magnetic resonance im- reduced in all 3 nuclei; differences in relative reduction aging (MRI) data have suggested volume reductions in did not differ among the nuclei. The remainder of the the mediodorsal (MDN) and pulvinar nuclei (PUL) of the thalamic volume (whole thalamus minus the volume of thalamus. The centromedian nucleus (CMN), impor- the 3 delineated nuclei) was not different between schizo- tant in attention and arousal, has not been previously stud- phrenic patients and controls, indicating that the vol- ied with MRI. ume reduction was specific to these nuclei. Volume rela- tive to brain size was reduced in all 3 nuclei and remained Methods: A sample of 41 patients with schizophrenia significant when only patients who had never been ex- (32 men and 9 women) and 60 healthy volunteers (45 posed to neuroleptic medication (n=15) were consid- men and 15 women) underwent assessment with high- ered. For the MDN, women had larger relative volumes resolution 1.2-mm thick anatomical MRI. Images were than men among controls, but men had larger volumes differentiated to enhance the edges and outline of the than women among schizophrenic patients. whole thalamus, and the MDN, PUL, and CMN were out- lined on all slices by a tracer masked to diagnostic Conclusions: Three association regions of the thala- status. -

Fmri Assessment of the Pulvinar and Medial Dorsal Nucleus in Normal Volunteers Monte S
Neuroscience Letters 404 (2006) 282–287 Thalamocortical circuits: fMRI assessment of the pulvinar and medial dorsal nucleus in normal volunteers Monte S. Buchsbaum a,∗, Bradley R. Buchsbaum b, Sylvie Chokron c,d, Cheuk Tang a,e, Tse-Chung Wei a, William Byne a,f a Department of Psychiatry, Mount Sinai School of Medicine, Box 1505, New York, NY 10029-6574, USA b Unit on Integrative Neuroimaging, Clinical Brain Disorders Branch, NIMH, NIH, Bethesda, MD, USA c Laboratoire de Psychologie Experimentale, CNRS, UMR 5105, Grenoble, France d Service de Neurologie, Fondation Ophtalmologique Rothschild, Paris, France e Department of Radiology, Mount Sinai School of Medicine, New York, NY, USA f Bronx VA Medical Center, Bronx, NY, USA Received 27 September 2005; received in revised form 10 March 2006; accepted 15 May 2006 Abstract This fMRI study investigates the activation of the thalamic nuclei in a spatial focusing-of-attention task previously shown to activate the pulvinar with FDG-PET and assesses the connectivity of the thalamic nuclei with cortical areas. Normal right-handed subjects (eight men, eight women, average age = 32 years) viewed four types of stimuli positioned to the right or left of the central fixation point (left hemifield-large letter, left hemifield-small letter display with flanking letters; right hemifield-large letter, right hemifield-small letter display with flankers). BOLD responses to small letters surrounded by flankers were compared with responses to large isolated letters. To examine maximum functional regional connectivity, we modeled “subject” as a random effect and attained fixed effect parameter estimates and t-statistics for functional connectivity between each of the thalamic nuclei (pulvinar, medial dorsal, and anterior) as the seed region and each non-seed voxel. -

Targeting the Centromedian Thalamic Nucleus for Deep Brain Stimulation
medRxiv preprint doi: https://doi.org/10.1101/19008136; this version posted October 18, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. It is made available under a CC-BY-NC-ND 4.0 International license . Title: Targeting the centromedian thalamic nucleus for deep brain stimulation Authors: Aaron E.L Warren1,2,3, Linda J. Dalic1,3,4, Wesley Thevathasan4,5,6, Annie Roten1, Kristian J. Bulluss5,7,8, John S. Archer1,2,3,4 1Department of Medicine (Austin Health), University of Melbourne, Heidelberg, Victoria, Australia 2Murdoch Children’s Research Institute, Parkville, Victoria, Australia 3The Florey Institute of Neuroscience and Mental Health, Heidelberg, Victoria, Australia 4Department of Neurology, Austin Health, Heidelberg, Victoria, Australia 5Bionics Institute, East Melbourne, Victoria, Australia 6Department of Medicine, University of Melbourne, and Department of Neurology, The Royal Melbourne Hospital, Parkville, Victoria, Australia 7Department of Neurosurgery, Austin Health, Heidelberg, Victoria, Australia 8Department of Surgery, University of Melbourne, Parkville, Victoria, Australia Corresponding author: Aaron E.L. Warren Ph: +613 9035 7110 Email: [email protected] Address: Melbourne Brain Centre, 245 Burgundy Street, Heidelberg, VIC, Australia 3084 Key words: Deep brain stimulation, centromedian nucleus, thalamus, epilepsy, Lennox- Gastaut syndrome, microelectrode recording, fMRI 1 NOTE: This preprint reports new research that has not been certified by peer review and should not be used to guide clinical practice. medRxiv preprint doi: https://doi.org/10.1101/19008136; this version posted October 18, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. -

Pulvinar Contributions to Visual Cortical Processing in the Rat
UC Irvine UC Irvine Electronic Theses and Dissertations Title Pulvinar contributions to visual cortical processing in the rat Permalink https://escholarship.org/uc/item/5xb9g4mp Author Scholl, Leo Russell Publication Date 2019 License https://creativecommons.org/licenses/by-nc/4.0/ 4.0 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA, IRVINE Pulvinar contributions to visual cortical processing in the rat DISSERTATION submitted in partial satisfaction of the requirements for the degree of DOCTOR OF PHILOSOPHY in Psychology by Leo Russell Scholl Dissertation Committee: Associate Professor David C. Lyon, Chair Professor Jeffrey L. Krichmar Distinguished Professor George Sperling 2019 c 2019 Leo Russell Scholl DEDICATION To the rats. ii TABLE OF CONTENTS Page LIST OF FIGURES v LIST OF TABLES vi ACKNOWLEDGMENTS vii CURRICULUM VITAE viii ABSTRACT OF THE DISSERTATION ix 1 Introduction 1 1.1 Background . 1 1.2 Open questions . 2 1.3 Specific aims . 5 1.4 References . 6 2 Characterizing visual responses to grating stimuli in the rat pulvinar nu- cleus 10 2.1 Abstract . 10 2.2 Introduction . 10 2.3 Methods . 15 2.4 Results . 20 2.5 Discussion . 29 2.6 References . 33 3 Projections between visual cortex and pulvinar nucleus in the rat 39 3.1 Abstract . 39 3.2 Introduction . 39 3.3 Methods . 42 3.4 Results . 46 3.5 Discussion . 53 3.6 References . 59 4 Thalamocortical modulation identified in vivo using new rabies virus vari- ant for bi-directional optical control 64 4.1 Abstract . 64 4.2 Introduction . -

Dual Midbrain and Forebrain Origins of Thalamic Inhibitory
RESEARCH ARTICLE Dual midbrain and forebrain origins of thalamic inhibitory interneurons Polona Jager1, Gerald Moore2,3, Padraic Calpin4, Xhuljana Durmishi1, Irene Salgarella1, Lucy Menage1, Yoshiaki Kita5, Yan Wang5, Dong Won Kim6, Seth Blackshaw6, Simon R Schultz2, Stephen Brickley3, Tomomi Shimogori5, Alessio Delogu1* 1Department of Basic and Clinical Neuroscience, Institute of Psychiatry, Psychology and Neuroscience, King’s College London, London, United Kingdom; 2Department of Bioengineering, Imperial College London, London, United Kingdom; 3Department of Life Sciences and Centre for Neurotechnology, Imperial College London, London, United Kingdom; 4Department of Physics and Astronomy, University College London, London, United Kingdom; 5RIKEN, Center for Brain Science (CBS), Saitama, Japan; 6The Solomon H. Snyder Department of Neuroscience, School of Medicine, Johns Hopkins University, Baltimore, United States Abstract The ubiquitous presence of inhibitory interneurons in the thalamus of primates contrasts with the sparsity of interneurons reported in mice. Here, we identify a larger than expected complexity and distribution of interneurons across the mouse thalamus, where all thalamic interneurons can be traced back to two developmental programmes: one specified in the midbrain and the other in the forebrain. Interneurons migrate to functionally distinct thalamocortical nuclei depending on their origin: the abundant, midbrain-derived class populates the first and higher order sensory thalamus while the rarer, forebrain-generated class is restricted to some higher order associative regions. We also observe that markers for the midbrain-born class are abundantly expressed throughout the thalamus of the New World monkey marmoset. These data therefore reveal that, despite the broad variability in interneuron density across mammalian *For correspondence: species, the blueprint of the ontogenetic organisation of thalamic interneurons of larger-brained [email protected] mammals exists and can be studied in mice. -

The Medial Division of the Medial Geniculate Body of the Cat: Implications for Thalamic Organization’
0270.6474/83/0312-2629$02.00/O The Journal of Neuroscience Copyright 0 Society for Neuroscience Vol. 3, No. 12, pp. 2629-2651 Printed in U.S.A. December 1983 THE MEDIAL DIVISION OF THE MEDIAL GENICULATE BODY OF THE CAT: IMPLICATIONS FOR THALAMIC ORGANIZATION’ JEFFERY A. WINER*,’ AND D. KENT MOREST$ * Department of Physiology-Anatomy, University of California, Berkeley, California 94720 and $ Department of Anatomy, The University of Connecticut Health Center, Farmington, Connecticut 06032 Received September 15,1982; Revised July 29, 1983; Accepted August 2,1983 Abstract The structure of neurons and axons was studied in the medial division of the medial geniculate body of the cat with the Golgi methods. The results show that the medial division consists of morphologically heterogeneous neurons. The main types, in descending order of frequency, are medium-sized neurons with (1) radiate, (2) tufted, or (3) elongate dendrites; (4) small stellate or radiate neurons, including Golgi type II cells with a locally arborizing, sparsely branching axon collateral system; (5) large neurons, which are weakly tufted. A variety of afferent axons impose a reticulate appearance on the fiber architecture of the medial division. The dominant element. in the neuropil consists of axons terminating in the medial division as well as a collateral system of fibers traveling to the adjacent ventral and dorsal divisions of the medial geniculate body. Four types of extrinsic axons are described, including two kinds of thin axons with collateral systems, thick fibers with restricted branches, and large axons with elaborate, serpentine collaterals. Compared to the dorsal and ventral divisions of the medial geniculate body, where, respectively, radiate and tufted neurons are more frequent, the medial division is intermediate in a sense-not that the degree of radiate or tufted dendritic branching is less well developed, but neither type of cell predominates. -

Thalamic Neuromodulation and Its Implications for Executive Networks
Thalamic neuromodulation and its implications for executive networks The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation Varela, Carmen. “Thalamic Neuromodulation and Its Implications for Executive Networks.” Front. Neural Circuits 8 (June 24, 2014). As Published http://dx.doi.org/10.3389/fncir.2014.00069 Publisher Frontiers Research Foundation Version Final published version Citable link http://hdl.handle.net/1721.1/89204 Terms of Use Creative Commons Attribution Detailed Terms http://creativecommons.org/licenses/by/4.0/ REVIEW ARTICLE published: 24 June 2014 doi: 10.3389/fncir.2014.00069 Thalamic neuromodulation and its implications for executive networks Carmen Varela* Picower Institute for Learning and Memory, Massachusetts Institute of Technology, Cambridge, MA, USA Edited by: The thalamus is a key structure that controls the routing of information in the brain. Guillermo Gonzalez-Burgos, Understanding modulation at the thalamic level is critical to understanding the flow of University of Pittsburgh, USA information to brain regions involved in cognitive functions, such as the neocortex, the Reviewed by: hippocampus, and the basal ganglia. Modulators contribute the majority of synapses Robert P.Vertes, Florida Atlantic University, USA that thalamic cells receive, and the highest fraction of modulator synapses is found in Randy M. Bruno, Columbia University, thalamic nuclei interconnected with higher order cortical regions. In addition, disruption USA of modulators often translates into disabling disorders of executive behavior. However, *Correspondence: modulation in thalamic nuclei such as the midline and intralaminar groups, which are Carmen Varela, Picower Institute for interconnected with forebrain executive regions, has received little attention compared Learning and Memory, Massachusetts Institute of Technology, Building 46, to sensory nuclei. -

Continuity Between Koniocellular Layers of Dorsal Lateral Geniculate and Inferior Pulvinar Nuclei in Common Marmosets
bioRxiv preprint doi: https://doi.org/10.1101/315598; this version posted July 3, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Continuity between koniocellular layers of dorsal lateral geniculate and inferior pulvinar nuclei in common marmosets Bing-Xing Huo*1, Natalie Zeater*2,3,4, Meng Kuan Lin1, Yeonsook S. Takahashi1, Mitsutoshi Hanada1, Jaimi Nagashima1, Brian C. Lee5, Junichi Hata1, Ulrike Grünert2,3,4, Michael I. Miller5, Marcello G.P. Rosa6,7, Hideyuki Okano1,8, Paul R. Martin2,3,4, Partha P. Mitra1,9 1 Laboratory for Marmoset Neural Architecture, RIKEN Center for Brain Science, Wako, Saitama, Japan 2 Save Sight Institute and Department of Clinical Ophthalmology, The University of Sydney, Sydney, NSW 2000, Australia 3Australian Research Council Centre of Excellence for Integrative Brain Function, The University of Sydney, Sydney, NSW 2000, Australia 4 School of Medical Sciences, The University of Sydney, Sydney, NSW 2000, Australia 5 Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD, USA 6 Department of Physiology and Biomedicine Research Institute, Monash University, Clayton, 3800 Victoria, Australia 7 Australian Research Council Centre of Excellence for Integrative Brain Function, Monash University Node, Clayton, 3800 Victoria, Australia 8 Department of Physiology, Keio University School of Medicine, Tokyo, Japan 9 Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 11724, USA -
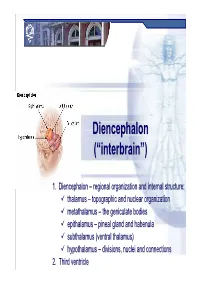
Diencephalondiencephalon ((““Interbraininterbrain ””))
DiencephalonDiencephalon ((““interbraininterbrain ””)) 1. Diencephalon – regional organization and internal structure: thalamus – topographic and nuclear organization metathalamus – the geniculate bodies epithalamus – pineal gland and habenula subthalamus (ventral thalamus) hypothalamus – divisions, nuclei and connections 2. Third ventricle Diencephalon EmbryologicEmbryologic developmentdevelopment Embryonic origin:origin side walls of the prosencephalon (forebrain) Location – at the midline of the brain: caudally – mesencephalon cranially – telencephalon Functions: relay system between sensory input neurons and other parts of the brain works in tandem with the limbic system Prof. Dr. Nikolai Lazarov 2 Diencephalon DiencephalonDiencephalon –– grossgross structurestructure andand partsparts Prof. Dr. Nikolai Lazarov 3 Thalamus ThalamusThalamus –– externalexternal featuresfeatures two egg -shaped lobes of grey matter Gr. θάλαµος = room , chamber third ventricle medially Gr. θάλαµος = room , chamber hypothalamus hypothalamic sulcus Thalamus dorsalis : nuclear complex – 2% of the total brain rostral pole = tuberculum anterius nuclear complex – 2% of the total brain thalami about 80% caudal pole = pulvinar thalami of diencephalic mass (“cushioned seat ”) ~30 mm long interthalamic adhesion ~20 mm wide lamina affixa ~20 mm tall stria terminalis thalami Prof. Dr. Nikolai Lazarov 4 Thalamus ThalamusThalamus –– internalinternal structurestructure internal medullary lamina three major nuclear masses: (medial) – Y-shaped