Cross-Amplification and Characterization of Microsatellite
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Fungal Interactions with Vascular and Non-Vascular Plants: an Investigation of Mutualisms and Their Roles in Heathland Regeneration
Fungal interactions with vascular and non-vascular plants: an investigation of mutualisms and their roles in heathland regeneration JILL KOWAL A THESIS SUBMITTED FOR THE DEGREE OF DOCTOR OF PHILOSOPHY, IMPERIAL COLLEGE LONDON, DEPARTMENT OF LIFE SCIENCES JUNE 16, 2016 1 Abstract Mycorrhizal mutualisms between aboveground vascular plant communities, which reward their belowground fungal associates with photosynthates in return for growth- limiting nutrients such as phosphate, are widely recognized as stable long-term interactions which helped plants colonize land. Pezoloma ericae (D.J. Read) Baral, an ascomycete mycorrhiza-forming fungus present amongst plants in the Ericales, such as heathers, also forms associations in several families of non-vascular leafy liverworts. Whether there is a mutually beneficial functional relationship between these leafy liverworts and the fungus growing in their rhizoids was previously unconfirmed. Furthermore, an ecological role of this ‘shared’ mycobiont and its link between vascular (Ericaceae) and non-vascular (liverworts) plants was also unknown. Thus the main questions asked in this dissertation are: 1) Is there a measurable mutually beneficial relationship between a liverwort and its fungal partner?; and, 2) Can liverworts harbouring the ericoid mycorrhiza P. ericae act as inoculum that facilitates the re- establishment of Ericaceae - and henceforth be proposed as a practical tool in a restoration ecology context. This is the first time British species of leafy liverworts are conclusively identified to harbour the ericoid mycorrhizal fungus Pezoloma ericae using molecular identification. I have demonstrated a mutualism occurring between the leafy liverworts and their fungal symbiont in two independent microcosm growth experiments and confirmatory reciprocal trophic exchanges between phosphorus and carbon and the two organisms. -

Diversity of Fungal Assemblages in Roots of Ericaceae in Two
Diversity of fungal assemblages in roots of Ericaceae in two Mediterranean contrasting ecosystems Ahlam Hamim, Lucie Miche, Ahmed Douaik, Rachid Mrabet, Ahmed Ouhammou, Robin Duponnois, Mohamed Hafidi To cite this version: Ahlam Hamim, Lucie Miche, Ahmed Douaik, Rachid Mrabet, Ahmed Ouhammou, et al.. Diversity of fungal assemblages in roots of Ericaceae in two Mediterranean contrasting ecosystems. Comptes Rendus Biologies, Elsevier Masson, 2017, 340 (4), pp.226-237. 10.1016/j.crvi.2017.02.003. hal- 01681523 HAL Id: hal-01681523 https://hal.archives-ouvertes.fr/hal-01681523 Submitted on 23 Apr 2018 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/315062117 Diversity of fungal assemblages in roots of Ericaceae in two Mediterranean contrasting ecosystems Article in Comptes rendus biologies · March 2017 DOI: 10.1016/j.crvi.2017.02.003 CITATIONS READS 0 37 7 authors, including: Ahmed Douaik Rachid Mrabet Institut National de Recherche Agronomique -

Ethnobotanical Study Among Ethnic Groups in the Shuiluo Valley, Southwest China: Local Knowledge of Ritual Plant Use
Ethnobotanical Study among Ethnic Groups in the Shuiluo Valley, Southwest China: Local Knowledge of Ritual Plant Use Master Thesis by Franziska Büeler 03-701-398 Submitted to Prof. Dr. Ulrike Müller-Böker Tutored by Dr. Caroline Weckerle and Prof. Dr. Ulrike Müller-Böker GEO 511 Master Thesis Department of Geography, University of Zurich October 2010 Contact: Franziska Büeler [email protected] Department of Human Geography, University Zurich Cover picture: Pumi woman holding the morning ritual on a flat roof, Siweng, Sichuan, China (Büeler 2010). TABLE OF CONTENTS FIGURES , TABLES , AND PHOTOGRAPHS ......................................................................... III ABSTRACT ................................................................................................................... V ACKNOWLEDGEMENTS ................................................................................................. VI GLOSSARY ................................................................................................................. VII 1 INTRODUCTION ...................................................................................................... 1 1.1 Framework of the study ............................................................................................................... 1 1.2 Research Objectives and Questions .......................................................................................... 2 2 THEORETICAL CONCEPTS ....................................................................................... 3 2.1 -

12. RHODODENDRON Linnaeus, Sp. Pl. 1: 392. 1753
Flora of China 14: 260–455. 2005. 12. RHODODENDRON Linnaeus, Sp. Pl. 1: 392. 1753. 杜鹃属 du juan shu Fang Mingyuan (方明渊), Fang Ruizheng (方瑞征 Fang Rhui-cheng), He Mingyou (何明友), Hu Linzhen (胡琳贞 Hu Ling-cheng), Yang Hanbi (杨汉碧 Yang Han-pi); David F. Chamberlain Shrubs or trees, terrestrial or epiphytic, with various hairs, and/or with peltate scales or glabrous, indumentum sometimes detersile (the hairs tangled and coming away as a layer). Leaves evergreen, deciduous or semideciduous, alternate, sometimes clustered at stem apex; margin entire, very rarely crenulate, abaxial indumentum sometimes with a pellicle (a thin skinlike layer on the surface). Inflorescence a raceme or corymb, mostly terminal, sometimes lateral, few- to many-flowered, sometimes reduced to a single flower. Calyx persistent, 5–8-lobed, sometimes reduced to a rim, lobes minute and triangular to large and conspicuous. Corolla funnelform, campanulate, tubular, rotate or hypocrateriform, regular or slightly zygomorphic, 5(–8)-lobed, lobes imbricate in bud. Stamens 5–10(–27), inserted at base of corolla, usually declinate; filaments linear to filiform, glabrous or pilose towards base; anthers without appendages, opening by terminal or oblique pores. Disk usually thick, 5–10(–14)-lobed. Ovary 5(–18)-locular, with hairs and/or scales, rarely glabrous. Style straight or declinate to deflexed, persistent; stigma capitate-discoid, crenate to lobed. Capsule cylindrical, coniform, or ovoid, sometimes curved, dehiscent from top, septicidal; valves thick or thin, straight or twisted. Seeds very numerous, minute, fusiform, always winged, or both ends with appendages or thread-like tails. About 1000 species: Asia, Europe, North America, two species in Australia; 571 species (409 endemic) in China. -

The Red List of Rhododendrons
The Red List of Rhododendrons Douglas Gibbs, David Chamberlain and George Argent BOTANIC GARDENS CONSERVATION INTERNATIONAL (BGCI) is a membership organization linking botanic gardens in over 100 countries in a shared commitment to biodiversity conservation, sustainable use and environmental education. BGCI aims to mobilize botanic gardens and work with partners to secure plant diversity for the well-being of people and the planet. BGCI provides the Secretariat for the IUCN/SSC Global Tree Specialist Group. Published by Botanic Gardens Conservation FAUNA & FLORA INTERNATIONAL (FFI) , founded in 1903 and the International, Richmond, UK world’s oldest international conservation organization, acts to conserve © 2011 Botanic Gardens Conservation International threatened species and ecosystems worldwide, choosing solutions that are sustainable, are based on sound science and take account of ISBN: 978-1-905164-35-6 human needs. Reproduction of any part of the publication for educational, conservation and other non-profit purposes is authorized without prior permission from the copyright holder, provided that the source is fully acknowledged. Reproduction for resale or other commercial purposes is prohibited without prior written permission from the copyright holder. THE GLOBAL TREES CAMPAIGN is undertaken through a partnership between FFI and BGCI, working with a wide range of other The designation of geographical entities in this document and the presentation of the material do not organizations around the world, to save the world’s most threatened trees imply any expression on the part of the authors and the habitats in which they grow through the provision of information, or Botanic Gardens Conservation International delivery of conservation action and support for sustainable use. -

Article Is Available Online Sponses to Climate, Soil and Plant Growth Form, Ecography, 36, At
Biogeosciences, 15, 2033–2053, 2018 https://doi.org/10.5194/bg-15-2033-2018 © Author(s) 2018. This work is distributed under the Creative Commons Attribution 4.0 License. Shrub type dominates the vertical distribution of leaf C : N : P stoichiometry across an extensive altitudinal gradient Wenqiang Zhao1, Peter B. Reich2, Qiannan Yu1,3, Ning Zhao4, Chunying Yin1, Chunzhang Zhao1, Dandan Li1, Jun Hu1, Ting Li1, Huajun Yin1, and Qing Liu1 1CAS Key Laboratory of Mountain Ecological Restoration and Bioresource Utilization & Ecological Restoration Biodiversity Conservation Key Laboratory of Sichuan Province, Chengdu Institute of Biology, Chinese Academy of Sciences, Chengdu 610041, China 2Department of Forest Resources and Institute on the Environment, University of Minnesota, Minnesota 55108, USA 3Southwest Jiaotong University & Faculty of Geosciences and Environmental Engineering, Chengdu 611756, China 4Cold and Arid Regions Environmental and Engineering Research Institute, Chinese Academy of Sciences, Lanzhou 730000, China Correspondence: Qing Liu ([email protected]) and Huajun Yin ([email protected]) Received: 14 November 2017 – Discussion started: 14 December 2017 Revised: 26 February 2018 – Accepted: 7 March 2018 – Published: 9 April 2018 Abstract. Understanding leaf stoichiometric patterns is cru- deciduous, broadleaf, and conifer), their leaf element con- cial for improving predictions of plant responses to environ- tents and responses to environments were remarkably differ- mental changes. Leaf stoichiometry of terrestrial ecosystems ent. Shrub type was the largest contributor to the total varia- has been widely investigated along latitudinal and longitudi- tions in leaf stoichiometry, while climate indirectly affected nal gradients. However, very little is known about the vertical the leaf C : N : P via its interactive effects on shrub type or distribution of leaf C : N : P and the relative effects of envi- soil. -
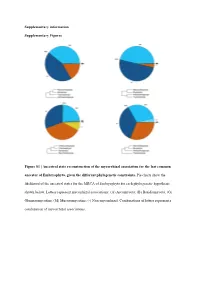
Ancestral State Reconstruction of the Mycorrhizal Association for the Last Common Ancestor of Embryophyta, Given the Different Phylogenetic Constraints
Supplementary information Supplementary Figures Figure S1 | Ancestral state reconstruction of the mycorrhizal association for the last common ancestor of Embryophyta, given the different phylogenetic constraints. Pie charts show the likelihood of the ancestral states for the MRCA of Embryophyta for each phylogenetic hypothesis shown below. Letters represent mycorrhizal associations: (A) Ascomycota; (B) Basidiomycota; (G) Glomeromycotina; (M) Mucoromycotina; (-) Non-mycorrhizal. Combinations of letters represent a combination of mycorrhizal associations. Austrocedrus chilensis Chamaecyparis obtusa Sequoiadendron giganteum Prumnopitys taxifolia Prumnopitys Prumnopitys montana Prumnopitys Prumnopitys ferruginea Prumnopitys Araucaria angustifolia Araucaria Dacrycarpus dacrydioides Dacrycarpus Taxus baccata Podocarpus oleifolius Podocarpus Afrocarpus falcatus Afrocarpus Ephedra fragilis Nymphaea alba Nymphaea Gnetum gnemon Abies alba Abies balsamea Austrobaileya scandens Austrobaileya Abies nordmanniana Thalictrum minus Thalictrum Abies homolepis Caltha palustris Caltha Abies magnifica ia repens Ranunculus Abies religiosa Ranunculus montanus Ranunculus Clematis vitalba Clematis Keteleeria davidiana Anemone patens Anemone Tsuga canadensis Vitis vinifera Vitis Tsuga mertensiana Saxifraga oppositifolia Saxifraga Larix decidua Hypericum maculatum Hypericum Larix gmelinii Phyllanthus calycinus Phyllanthus Larix kaempferi Hieronyma oblonga Hieronyma Pseudotsuga menziesii Salix reinii Salix Picea abies Salix polaris Salix Picea crassifolia Salix herbacea -

Evergreen Sclerophyllous Quercus Forests in Northwestern Yunnan
Web Ecology 6: 88–101. Evergreen sclerophyllous Quercus forests in northwestern Yunnan, China as compared to the Mediterranean evergreen Quercus forests in California, USA and northeastern Spain C. Q. Tang Tang, C. Q. 2006. Evergreen sclerophyllous Quercus forests in northwestern Yunnan, China as compared to the Mediterranean evergreen Quercus forests in California, USA and northeastern Spain. – Web Ecol. 6: 88–101. Evergreen sclerophyllous Quercus forests in NW Yunnan (China) were studied and compared with the Mediterranean evergreen sclerophyllous Quercus forests in central coastal California (USA) and Catalonia (NE Spain). Forests of Q. aquifolioides, Q. pan- nosa, Q. longispica of NW Yunnan, Q. agrifolia of California and Q. ilex of NE Spain were analyzed as representative communities. The similarities and differences at the community level in the contemporary vegetation of the sclerophyllous Quercus forest found in the three regions are clarified. The general patterns of the evergreen Quercus forest in the three regions were similar, though different assemblages of species were involved. The species diversity in all three regions was rather low. The species richness did not significantly differ among the forests, although in the Q. longispica forest it is somewhat higher than the others. The three representative species of evergreen Quercus in NW Yunnan reached the greatest maximum height, while Q. agrifolia of California had the largest basal area per ha. The Q. ilex forest of Spain had the lowest values for maximum tree height and dbh and the highest density per ha. Frequency of dbh size classes indicated that Q. aquifolioides, Q. pannosa, and Q. agrifolia had potentially good regeneration of the sporadic type with highest values for the intermediate size classes, and the regeneration of Q. -

Seifertia Shangrilaensis Sp. Nov. (Melanommataceae), a New Species from Southwest China
Phytotaxa 273 (1): 034–042 ISSN 1179-3155 (print edition) http://www.mapress.com/j/pt/ PHYTOTAXA Copyright © 2016 Magnolia Press Article ISSN 1179-3163 (online edition) http://dx.doi.org/10.11646/phytotaxa.273.1.3 Seifertia shangrilaensis sp. nov. (Melanommataceae), a new species from Southwest China JUNFU LI1,2,3,4,7, RUNGTIWA PHOOKAMSAK2,3,4,7, AUSANA MAPOOK3,4, SARANYAPHAT BOONMEE3, JARAYAMA D. BHAT5,6, KEVIN D. HYDE1,2,3,4 & SAISAMORN LUMYONG1,* 1Department of Biology, Faculty of Science, Chinag Mai University, Chiang Mai 50200, Thailand. 2World Agroforestry Centre, East and Central Asia, 132 Lanhei Road, Kunming 650201, China. 3Center of Excellence in Fungal Research, Mae Fah Luang University, Chiang Rai 57100, Thailand. 4School of Science, Mae Fah Luang University, Chiang Rai 57100, Thailand. 5Formerly, Department of Botany, Goa University, Goa-403206, India. 6128/1-J, Azad Housing Society, Curca, Goa Velha-403108, India. 7Key Laboratory of Biodiversity and Biogeography, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, China. Correspondence to*: [email protected] Abstract A new Seifertia species was isolated from hanging rachides of Rhododendron decorum in Yunnan Province, Southwest China. The new taxon was compared with the type species, S. azalea and differs in having wider conidiophores, with hya- line to subhyaline and smaller conidia, while S. azalea has olive-brown to brown, rarely branched conidiophores, and pale brown or olive-brown, very rarely septate conidia. Phylogenetic analyses of combined LSU, SSU and TEF1-α sequence data show that S. shangrilaensis forms a robust clade with S. azalea nested among the species of Melanommataceae in the order Pleosporales. -

The International Rhododendron Register & Checklist Consolidated Supplement
THE ROYAL HORTICULTURAL SOCIETY The International RHODODENDRON REGISTER and CHECKLIST (2004) Consolidated Supplement 2003-2017 THE INTERNATIONAL RHODODENDRON REGISTER AND CHECKLIST (2004) Consolidated Supplement 2003-2017 The second edition of the International Rhododendron Register and Checklist included accounts of all plants whose names had been registered up to the end of December 2002. This Supplement deals with all names registered during the period 1 January 2003 to 31 December 2017. The layout of each entry follows the format adopted in the 2004 Register and Checklist and is explained there in detail on pages 7-10 of the Introduction. It may however be helpful to repeat here that the following abbreviations are employed: (a) = azalea (az) = azaleodendron cv. = cultivar G = grown to first flower by… gp = Group H = hybridised by… I = introduced by… N = named by… (r) = rhododendron R = raised by… REG = registered by… S = selected by… (s) = seed parent (v) = vireya Wherever possible colour references are given to the RHS Colour Chart (1966, 1986, 1995 & 2001) with the colour description being taken from A contribution towards standardization of color names in horticulture, by R.D. Huse and K.L. Kelly (ed. D.H. Voss), 1984. All raisers and introducers of new rhododendrons are urged to ensure that the names of their plants have been registered and are reminded that registration needs to be effected before the plant is introduced or the name appears in print. Registration forms can be obtained from the International Rhododendron Registrar (at the address given below), from the RHS website (www.rhs.org.uk/plants/plant-science/plant registration-forms/rhodoform) or from one of the national or regional registrars as follows: North America Mr Michael Martin Mills, 632 Burnham Road, Philadelphia, Pennsylvania PA19119-3510, USA Australia Mrs Lesley Eaton, PO Box 597, Snug, Tasmania 7054, Australia New Zealand Mr Brian J. -

Latin for Gardeners: Over 3,000 Plant Names Explained and Explored
L ATIN for GARDENERS ACANTHUS bear’s breeches Lorraine Harrison is the author of several books, including Inspiring Sussex Gardeners, The Shaker Book of the Garden, How to Read Gardens, and A Potted History of Vegetables: A Kitchen Cornucopia. The University of Chicago Press, Chicago 60637 © 2012 Quid Publishing Conceived, designed and produced by Quid Publishing Level 4, Sheridan House 114 Western Road Hove BN3 1DD England Designed by Lindsey Johns All rights reserved. Published 2012. Printed in China 22 21 20 19 18 17 16 15 14 13 1 2 3 4 5 ISBN-13: 978-0-226-00919-3 (cloth) ISBN-13: 978-0-226-00922-3 (e-book) Library of Congress Cataloging-in-Publication Data Harrison, Lorraine. Latin for gardeners : over 3,000 plant names explained and explored / Lorraine Harrison. pages ; cm ISBN 978-0-226-00919-3 (cloth : alkaline paper) — ISBN (invalid) 978-0-226-00922-3 (e-book) 1. Latin language—Etymology—Names—Dictionaries. 2. Latin language—Technical Latin—Dictionaries. 3. Plants—Nomenclature—Dictionaries—Latin. 4. Plants—History. I. Title. PA2387.H37 2012 580.1’4—dc23 2012020837 ∞ This paper meets the requirements of ANSI/NISO Z39.48-1992 (Permanence of Paper). L ATIN for GARDENERS Over 3,000 Plant Names Explained and Explored LORRAINE HARRISON The University of Chicago Press Contents Preface 6 How to Use This Book 8 A Short History of Botanical Latin 9 Jasminum, Botanical Latin for Beginners 10 jasmine (p. 116) An Introduction to the A–Z Listings 13 THE A-Z LISTINGS OF LatIN PlaNT NAMES A from a- to azureus 14 B from babylonicus to byzantinus 37 C from cacaliifolius to cytisoides 45 D from dactyliferus to dyerianum 69 E from e- to eyriesii 79 F from fabaceus to futilis 85 G from gaditanus to gymnocarpus 94 H from haastii to hystrix 102 I from ibericus to ixocarpus 109 J from jacobaeus to juvenilis 115 K from kamtschaticus to kurdicus 117 L from labiatus to lysimachioides 118 Tropaeolum majus, M from macedonicus to myrtifolius 129 nasturtium (p. -

Comparative Analysis of Microsatellite, SNP, and Indel Markers in Four Rhododendron Species Based on RNA-Seq
Breeding Science 68: 536–544 (2018) doi:10.1270/jsbbs.18092 Research Paper Comparative analysis of microsatellite, SNP, and InDel markers in four Rhododendron species based on RNA-seq Shuzhen Wang, Zhiliang Li, Xudong Guo, Yuanping Fang, Jun Xiang and Weibin Jin* Hubei Key Laboratory of Economic Forest Germplasm Improvement and Resources Comprehensive Utilization; Hubei Collaborative Innovation Center for the Characteristic Resources Exploitation of Dabie Mountains; College of Life Science, Huanggang Normal University, Huanggang, 438000, Hubei Province, P.R. China Rhododendron possesses valuable horticultural and medicinal properties. However, the genetic studies have been hindered due to the lack of genetic markers. Based on RNA-seq, large-scale molecular markers were de- veloped from four Rhododendron species endemic to Dabie Mountains (central China): R. fortunei (5.25 Gb; SSRs, 12,756, one/2.37 kb, 147 types; SNPs, 38,313; InDels, 3,174), R. simsii (5.80 Gb; SSRs, 13,294, one/2.58 kb, 167 types; SNPs, 136,590; InDels, 6,258), R. mariesii (6.53 Gb; SSRs, 15,724, one/2.51 kb, 170 types; SNPs, 44,942; InDels, 4,126), and R. molle (4.35 Gb; SSRs, 10,214, one/2.49 kb, 110 types; SNPs, 77,829; InDels, 3,416). Di-nucleotide repeats were the main type (59.126%–64.314%), and AG/CT repeat (55.18%–61.22%) was the most. In particular, 89 species-specific types had been found. Furthermore, C:G→T:A mutation was the main SNP type (30.475%–34.99%). However, C:G→G:C mutation was the least type in R.