Bayesian Hypothesis Tests Using Nonparametric Statistics
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Nonparametric Statistics for Social and Behavioral Sciences
Statistics Incorporating a hands-on approach, Nonparametric Statistics for Social SOCIAL AND BEHAVIORAL SCIENCES and Behavioral Sciences presents the concepts, principles, and methods NONPARAMETRIC STATISTICS FOR used in performing many nonparametric procedures. It also demonstrates practical applications of the most common nonparametric procedures using IBM’s SPSS software. Emphasizing sound research designs, appropriate statistical analyses, and NONPARAMETRIC accurate interpretations of results, the text: • Explains a conceptual framework for each statistical procedure STATISTICS FOR • Presents examples of relevant research problems, associated research questions, and hypotheses that precede each procedure • Details SPSS paths for conducting various analyses SOCIAL AND • Discusses the interpretations of statistical results and conclusions of the research BEHAVIORAL With minimal coverage of formulas, the book takes a nonmathematical ap- proach to nonparametric data analysis procedures and shows you how they are used in research contexts. Each chapter includes examples, exercises, SCIENCES and SPSS screen shots illustrating steps of the statistical procedures and re- sulting output. About the Author Dr. M. Kraska-Miller is a Mildred Cheshire Fraley Distinguished Professor of Research and Statistics in the Department of Educational Foundations, Leadership, and Technology at Auburn University, where she is also the Interim Director of Research for the Center for Disability Research and Service. Dr. Kraska-Miller is the author of four books on teaching and communications. She has published numerous articles in national and international refereed journals. Her research interests include statistical modeling and applications KRASKA-MILLER of statistics to theoretical concepts, such as motivation; satisfaction in jobs, services, income, and other areas; and needs assessments particularly applicable to special populations. -

Logrank Tests (Freedman)
PASS Sample Size Software NCSS.com Chapter 702 Logrank Tests (Freedman) Introduction This module allows the sample size and power of the logrank test to be analyzed under the assumption of proportional hazards. Time periods are not stated. Rather, it is assumed that enough time elapses to allow for a reasonable proportion of responses to occur. If you want to study the impact of accrual and follow-up time, you should use the one of the other logrank modules available in PASS. The formulas used in this module come from Machin et al. (2018). They are also given in Fayers and Machin (2016) where they are applied to sizing quality of life studies. They were originally published in Freedman (1982) and are often referred to by that name. A clinical trial is often employed to test the equality of survival distributions for two treatment groups. For example, a researcher might wish to determine if Beta-Blocker A enhances the survival of newly diagnosed myocardial infarction patients over that of the standard Beta-Blocker B. The question being considered is whether the pattern of survival is different. The two-sample t-test is not appropriate for two reasons. First, the data consist of the length of survival (time to failure), which is often highly skewed, so the usual normality assumption cannot be validated. Second, since the purpose of the treatment is to increase survival time, it is likely (and desirable) that some of the individuals in the study will survive longer than the planned duration of the study. The survival times of these individuals are then unobservable and are said to be censored. -

Randomization-Based Test for Censored Outcomes: a New Look at the Logrank Test
Randomization-based Test for Censored Outcomes: A New Look at the Logrank Test Xinran Li and Dylan S. Small ∗ Abstract Two-sample tests have been one of the most classical topics in statistics with wide appli- cation even in cutting edge applications. There are at least two modes of inference used to justify the two-sample tests. One is usual superpopulation inference assuming the units are independent and identically distributed (i.i.d.) samples from some superpopulation; the other is finite population inference that relies on the random assignments of units into different groups. When randomization is actually implemented, the latter has the advantage of avoiding distribu- tional assumptions on the outcomes. In this paper, we will focus on finite population inference for censored outcomes, which has been less explored in the literature. Moreover, we allow the censoring time to depend on treatment assignment, under which exact permutation inference is unachievable. We find that, surprisingly, the usual logrank test can also be justified by ran- domization. Specifically, under a Bernoulli randomized experiment with non-informative i.i.d. censoring within each treatment arm, the logrank test is asymptotically valid for testing Fisher's null hypothesis of no treatment effect on any unit. Moreover, the asymptotic validity of the lo- grank test does not require any distributional assumption on the potential event times. We further extend the theory to the stratified logrank test, which is useful for randomized blocked designs and when censoring mechanisms vary across strata. In sum, the developed theory for the logrank test from finite population inference supplements its classical theory from usual superpopulation inference, and helps provide a broader justification for the logrank test. -

Nonparametric Estimation by Convex Programming
The Annals of Statistics 2009, Vol. 37, No. 5A, 2278–2300 DOI: 10.1214/08-AOS654 c Institute of Mathematical Statistics, 2009 NONPARAMETRIC ESTIMATION BY CONVEX PROGRAMMING By Anatoli B. Juditsky and Arkadi S. Nemirovski1 Universit´eGrenoble I and Georgia Institute of Technology The problem we concentrate on is as follows: given (1) a convex compact set X in Rn, an affine mapping x 7→ A(x), a parametric family {pµ(·)} of probability densities and (2) N i.i.d. observations of the random variable ω, distributed with the density pA(x)(·) for some (unknown) x ∈ X, estimate the value gT x of a given linear form at x. For several families {pµ(·)} with no additional assumptions on X and A, we develop computationally efficient estimation routines which are minimax optimal, within an absolute constant factor. We then apply these routines to recovering x itself in the Euclidean norm. 1. Introduction. The problem we are interested in is essentially as fol- lows: suppose that we are given a convex compact set X in Rn, an affine mapping x A(x) and a parametric family pµ( ) of probability densities. Suppose that7→N i.i.d. observations of the random{ variable· } ω, distributed with the density p ( ) for some (unknown) x X, are available. Our objective A(x) · ∈ is to estimate the value gT x of a given linear form at x. In nonparametric statistics, there exists an immense literature on various versions of this problem (see, e.g., [10, 11, 12, 13, 15, 17, 18, 21, 22, 23, 24, 25, 26, 27, 28] and the references therein). -

Survival Analysis Using a 5‐Step Stratified Testing and Amalgamation
Received: 13 March 2020 Revised: 25 June 2020 Accepted: 24 August 2020 DOI: 10.1002/sim.8750 RESEARCH ARTICLE Survival analysis using a 5-step stratified testing and amalgamation routine (5-STAR) in randomized clinical trials Devan V. Mehrotra Rachel Marceau West Biostatistics and Research Decision Sciences, Merck & Co., Inc., North Wales, Randomized clinical trials are often designed to assess whether a test treatment Pennsylvania, USA prolongs survival relative to a control treatment. Increased patient heterogene- ity, while desirable for generalizability of results, can weaken the ability of Correspondence Devan V. Mehrotra, Biostatistics and common statistical approaches to detect treatment differences, potentially ham- Research Decision Sciences, Merck & Co., pering the regulatory approval of safe and efficacious therapies. A novel solution Inc.,NorthWales,PA,USA. Email: [email protected] to this problem is proposed. A list of baseline covariates that have the poten- tial to be prognostic for survival under either treatment is pre-specified in the analysis plan. At the analysis stage, using all observed survival times but blinded to patient-level treatment assignment, “noise” covariates are removed with elastic net Cox regression. The shortened covariate list is used by a condi- tional inference tree algorithm to segment the heterogeneous trial population into subpopulations of prognostically homogeneous patients (risk strata). After patient-level treatment unblinding, a treatment comparison is done within each formed risk stratum and stratum-level results are combined for overall statis- tical inference. The impressive power-boosting performance of our proposed 5-step stratified testing and amalgamation routine (5-STAR), relative to that of the logrank test and other common approaches that do not leverage inherently structured patient heterogeneity, is illustrated using a hypothetical and two real datasets along with simulation results. -

Repeated-Measures ANOVA & Friedman Test Using STATCAL
Repeated-Measures ANOVA & Friedman Test Using STATCAL (R) & SPSS Prana Ugiana Gio Download STATCAL in www.statcal.com i CONTENT 1.1 Example of Case 1.2 Explanation of Some Book About Repeated-Measures ANOVA 1.3 Repeated-Measures ANOVA & Friedman Test 1.4 Normality Assumption and Assumption of Equality of Variances (Sphericity) 1.5 Descriptive Statistics Based On SPSS dan STATCAL (R) 1.6 Normality Assumption Test Using Kolmogorov-Smirnov Test Based on SPSS & STATCAL (R) 1.7 Assumption Test of Equality of Variances Using Mauchly Test Based on SPSS & STATCAL (R) 1.8 Repeated-Measures ANOVA Based on SPSS & STATCAL (R) 1.9 Multiple Comparison Test Using Boferroni Test Based on SPSS & STATCAL (R) 1.10 Friedman Test Based on SPSS & STATCAL (R) 1.11 Multiple Comparison Test Using Wilcoxon Test Based on SPSS & STATCAL (R) ii 1.1 Example of Case For example given data of weight of 11 persons before and after consuming medicine of diet for one week, two weeks, three weeks and four week (Table 1.1.1). Tabel 1.1.1 Data of Weight of 11 Persons Weight Name Before One Week Two Weeks Three Weeks Four Weeks A 89.43 85.54 80.45 78.65 75.45 B 85.33 82.34 79.43 76.55 71.35 C 90.86 87.54 85.45 80.54 76.53 D 91.53 87.43 83.43 80.44 77.64 E 90.43 84.45 81.34 78.64 75.43 F 90.52 86.54 85.47 81.44 78.64 G 87.44 83.34 80.54 78.43 77.43 H 89.53 86.45 84.54 81.35 78.43 I 91.34 88.78 85.47 82.43 78.76 J 88.64 84.36 80.66 78.65 77.43 K 89.51 85.68 82.68 79.71 76.5 Average 89.51 85.68 82.68 79.71 76.69 Based on Table 1.1.1: The person whose name is A has initial weight 89,43, after consuming medicine of diet for one week 85,54, two weeks 80,45, three weeks 78,65 and four weeks 75,45. -

Applied Biostatistics Applied Biostatistics for the Pulmonologist
APPLIED BIOSTATISTICS FOR THE PULMONOLOGIST DR. VISHWANATH GELLA • Statistics is a way of thinking about the world and decision making-By Sir RA Fisher Why do we need statistics? • A man with one watch always knows what time it is • A man with two watches always searches to identify the correct one • A man with ten watches is always reminded of the difficu lty in measuri ng ti me Objectives • Overview of Biostatistical Terms and Concepts • Application of Statistical Tests Types of statistics • Descriptive Statistics • identify patterns • leads to hypothesis generation • Inferential Statistics • distinguish true differences from random variation • allows hypothesis testing Study design • Analytical studies Case control study(Effect to cause) Cohort study(Cause to effect) • Experimental studies Randomized controlled trials Non-randomized trials Sample size estimation • ‘Too small’ or ‘Too large’ • Sample size depends upon four critical quantities: Type I & II error stat es ( al ph a & b et a errors) , th e vari abilit y of th e d at a (S.D)2 and the effect size(d) • For two group parallel RCT with a continuous outcome - sample size(n) per group = 16(S.D)2/d2 for fixed alpha and beta values • Anti hypertensive trial- effect size= 5 mmHg, S.D of the data- 10 mm Hggp. n= 16 X 100/25= 64 patients per group in the study • Statistical packages - PASS in NCSS, n query or sample power TYPES OF DATA • Quant itat ive (“how muc h?”) or categor ica l variable(“what type?”) • QiiiblQuantitative variables 9 continuous- Blood pressure, height, weight or age 9 Discrete- No. -
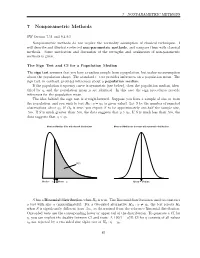
7 Nonparametric Methods
7 NONPARAMETRIC METHODS 7 Nonparametric Methods SW Section 7.11 and 9.4-9.5 Nonparametric methods do not require the normality assumption of classical techniques. I will describe and illustrate selected non-parametric methods, and compare them with classical methods. Some motivation and discussion of the strengths and weaknesses of non-parametric methods is given. The Sign Test and CI for a Population Median The sign test assumes that you have a random sample from a population, but makes no assumption about the population shape. The standard t−test provides inferences on a population mean. The sign test, in contrast, provides inferences about a population median. If the population frequency curve is symmetric (see below), then the population median, iden- tified by η, and the population mean µ are identical. In this case the sign procedures provide inferences for the population mean. The idea behind the sign test is straightforward. Suppose you have a sample of size m from the population, and you wish to test H0 : η = η0 (a given value). Let S be the number of sampled observations above η0. If H0 is true, you expect S to be approximately one-half the sample size, .5m. If S is much greater than .5m, the data suggests that η > η0. If S is much less than .5m, the data suggests that η < η0. Mean and Median differ with skewed distributions Mean and Median are the same with symmetric distributions 50% Median = η Mean = µ Mean = Median S has a Binomial distribution when H0 is true. The Binomial distribution is used to construct a test with size α (approximately). -

9 Blocked Designs
9 Blocked Designs 9.1 Friedman's Test 9.1.1 Application 1: Randomized Complete Block Designs • Assume there are k treatments of interest in an experiment. In Section 8, we considered the k-sample Extension of the Median Test and the Kruskal-Wallis Test to test for any differences in the k treatment medians. • Suppose the experimenter is still concerned with studying the effects of a single factor on a response of interest, but variability from another factor that is not of interest is expected. { Suppose a researcher wants to study the effect of 4 fertilizers on the yield of cotton. The researcher also knows that the soil conditions at the 8 areas for performing an experiment are highly variable. Thus, the researcher wants to design an experiment to detect any differences among the 4 fertilizers on the cotton yield in the presence a \nuisance variable" not of interest (the 8 areas). • Because experimental units can vary greatly with respect to physical characteristics that can also influence the response, the responses from experimental units that receive the same treatment can also vary greatly. • If it is not controlled or accounted for in the data analysis, it can can greatly inflate the experimental variability making it difficult to detect real differences among the k treatments of interest (large Type II error). • If this source of variability can be separated from the treatment effects and the random experimental error, then the sensitivity of the experiment to detect real differences between treatments in increased (i.e., lower the Type II error). • Therefore, the goal is to choose an experimental design in which it is possible to control the effects of a variable not of interest by bringing experimental units that are similar into a group called a \block". -

DETECTION MONITORING TESTS Unified Guidance
PART III. DETECTION MONITORING TESTS Unified Guidance PART III. DETECTION MONITORING TESTS This third part of the Unified Guidance presents core procedures recommended for formal detection monitoring at RCRA-regulated facilities. Chapter 16 describes two-sample tests appropriate for some small facilities, facilities in interim status, or for periodic updating of background data. These tests include two varieties of the t-test and two non-parametric versions-- the Wilcoxon rank-sum and Tarone-Ware procedures. Chapter 17 discusses one-way analysis of variance [ANOVA], tolerance limits, and the application of trend tests during detection monitoring. Chapter 18 is a primer on several kinds of prediction limits, which are combined with retesting strategies in Chapter 19 to address the statistical necessity of performing multiple comparisons during RCRA statistical evaluations. Retesting is also discussed in Chapter 20, which presents control charts as an alternative to prediction limits. As discussed in Section 7.5, any of these detection-level tests may also be applied to compliance/assessment and corrective action monitoring, where a background groundwater protection standard [GWPS] is defined as a critical limit using two- or multiple-sample comparison tests. Caveats and limitations discussed for detection monitoring tests are also relevant to this situation. To maintain continuity of presentation, this additional application is presumed but not repeated in the following specific test and procedure discussions. Although other users and programs may find these statistical tests of benefit due to their wider applicability to other environmental media and types of data, the methods described in Parts III and IV are primarily tailored to the RCRA setting and designed to address formal RCRA monitoring requirements. -

4. Comparison of Two (K) Samples K=2 Problem: Compare the Survival Distributions Between Two Groups
4. Comparison of Two (K) Samples K=2 Problem: compare the survival distributions between two groups. Ex: comparing treatments on patients with a particular disease. 푍: Treatment indicator, i.e. 푍 = 1 for treatment 1 (new treatment); 푍 = 0 for treatment 0 (standard treatment or placebo) Null Hypothesis: H0: no treatment (group) difference H0: 푆0 푡 = 푆1 푡 , for 푡 ≥ 0 H0: 휆0 푡 = 휆1 푡 , for 푡 ≥ 0 Alternative Hypothesis: Ha: the survival time for one treatment is stochastically larger or smaller than the survival time for the other treatment. Ha: 푆1 푡 ≥ 푆0 푡 , for 푡 ≥ 0 with strict inequality for some 푡 (one-sided) Ha: either 푆1 푡 ≥ 푆0 푡 , or 푆0 푡 ≥ 푆1 푡 , for 푡 ≥ 0 with strict inequality for some 푡 Solution: In biomedical applications, it has become common practice to use nonparametric tests; that is, using test statistics whose distribution under the null hypothesis does not depend on specific parametric assumptions on the shape of the probability distribution. With censored survival data, the class of weighted logrank tests are mostly used, with the logrank test being the most commonly used. Notations A sample of triplets 푋푖, Δ푖, 푍푖 , 푖 = 1, 2, … , 푛, where 1 푛푒푤 푡푟푒푎푡푚푒푛푡 푋푖 = min(푇푖, 퐶푖) Δ푖 = 퐼 푇푖 ≤ 퐶푖 푍 = ቊ 푖 0 푠푡푎푛푑푎푟푑 푇푟푒푎푡푚푒푛푡 푇푖 = latent failure time; 퐶푖 = latent censoring time Also, define, 푛 푛1 = number of individuals in group 1 푛푗 = 퐼(푍푗 = 푗) , 푗 = 0, 1 푛0 = number of individuals in group 0 푖=1 푛 = 푛0 + 푛1 푛 푌1(푥) = number of individuals at risk at time 푥 from trt 1 = σ푖=1 퐼(푋푖 ≥ 푥, 푍푖 = 1) 푛 푌0(푥) = number of individuals at risk at time 푥 from trt 0 = σ푖=1 퐼(푋푖 ≥ 푥, 푍푖 = 0) 푌(푥) = 푌0(푥) + 푌1(푥) 푛 푑푁1(푥) = # of deaths observed at time 푥 from trt 1 = σ푖=1 퐼(푋푖 = 푥, Δ푖 = 1, 푍푖 = 1) 푛 푑푁0(푥) = # of deaths observed at time 푥 from trt 0 = σ푖=1 퐼(푋푖 = 푥, Δ푖 = 1, 푍푖 = 0) 푛 푑푁 푥 = 푑푁0 푥 + 푑푁1 푥 = σ푖=1 퐼(푋푖 = 푥, Δ푖 = 1) Note: 푑푁 푥 actually correspond to the observed number of deaths in time window 푥, 푥 + Δ푥 for some partition of the time axis into intervals of length Δ푥. -

Kaplan-Meier Curves (Logrank Tests)
NCSS Statistical Software NCSS.com Chapter 555 Kaplan-Meier Curves (Logrank Tests) Introduction This procedure computes the nonparametric Kaplan-Meier and Nelson-Aalen estimates of survival and associated hazard rates. It can fit complete, right censored, left censored, interval censored (readout), and grouped data values. It outputs various statistics and graphs that are useful in reliability and survival analysis. It also performs several logrank tests and provides both the parametric and randomization test significance levels. This procedure also computes restricted mean survival time (RMST) and restricted mean time lost (RMTL) statistics and associated between-group comparisons. Overview of Survival Analysis We will give a brief introduction to the subject in this section. For a complete account of survival analysis, we suggest the book by Klein and Moeschberger (2003). Survival analysis is the study of the distribution of life times. That is, it is the study of the elapsed time between an initiating event (birth, start of treatment, diagnosis, or start of operation) and a terminal event (death, relapse, cure, or machine failure). The data values are a mixture of complete (terminal event occurred) and censored (terminal event has not occurred) observations. From the data values, the survival analyst makes statements about the survival distribution of the failure times. This distribution allows questions about such quantities as survivability, expected life time, and mean time to failure to be answered. Let T be the elapsed time until the occurrence of a specified event. The event may be death, occurrence of a disease, disappearance of a disease, appearance of a tumor, etc. The probability distribution of T may be specified using one of the following basic functions.