9 Blocked Designs
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

14. Non-Parametric Tests You Know the Story. You've Tried for Hours To
14. Non-Parametric Tests You know the story. You’ve tried for hours to normalise your data. But you can’t. And even if you can, your variances turn out to be unequal anyway. But do not dismay. There are statistical tests that you can run with non-normal, heteroscedastic data. All of the tests that we have seen so far (e.g. t-tests, ANOVA, etc.) rely on comparisons with some probability distribution such as a t-distribution or f-distribution. These are known as “parametric statistics” as they make inferences about population parameters. However, there are other statistical tests that do not have to meet the same assumptions, known as “non-parametric tests.” Many of these tests are based on ranks and/or are comparisons of sample medians rather than means (as means of non-normal data are not meaningful). Non-parametric tests are often not as powerful as parametric tests, so you may find that your p-values are slightly larger. However, if your data is highly non-normal, non-parametric test may detect an effect that is missed in the equivalent parametric test due to falsely large variances caused by the skew of the data. There is, of course, no problem in analyzing normally distributed data with non-parametric statistics also. Non-parametric tests are equally valid (i.e. equally defendable) and most have been around as long as their parametric counterparts. So, if your data does not meet the assumption of normality or homoscedasticity and if it is impossible (or inappropriate) to transform it, you can use non-parametric tests. -

Nonparametric Statistics for Social and Behavioral Sciences
Statistics Incorporating a hands-on approach, Nonparametric Statistics for Social SOCIAL AND BEHAVIORAL SCIENCES and Behavioral Sciences presents the concepts, principles, and methods NONPARAMETRIC STATISTICS FOR used in performing many nonparametric procedures. It also demonstrates practical applications of the most common nonparametric procedures using IBM’s SPSS software. Emphasizing sound research designs, appropriate statistical analyses, and NONPARAMETRIC accurate interpretations of results, the text: • Explains a conceptual framework for each statistical procedure STATISTICS FOR • Presents examples of relevant research problems, associated research questions, and hypotheses that precede each procedure • Details SPSS paths for conducting various analyses SOCIAL AND • Discusses the interpretations of statistical results and conclusions of the research BEHAVIORAL With minimal coverage of formulas, the book takes a nonmathematical ap- proach to nonparametric data analysis procedures and shows you how they are used in research contexts. Each chapter includes examples, exercises, SCIENCES and SPSS screen shots illustrating steps of the statistical procedures and re- sulting output. About the Author Dr. M. Kraska-Miller is a Mildred Cheshire Fraley Distinguished Professor of Research and Statistics in the Department of Educational Foundations, Leadership, and Technology at Auburn University, where she is also the Interim Director of Research for the Center for Disability Research and Service. Dr. Kraska-Miller is the author of four books on teaching and communications. She has published numerous articles in national and international refereed journals. Her research interests include statistical modeling and applications KRASKA-MILLER of statistics to theoretical concepts, such as motivation; satisfaction in jobs, services, income, and other areas; and needs assessments particularly applicable to special populations. -

Nonparametric Estimation by Convex Programming
The Annals of Statistics 2009, Vol. 37, No. 5A, 2278–2300 DOI: 10.1214/08-AOS654 c Institute of Mathematical Statistics, 2009 NONPARAMETRIC ESTIMATION BY CONVEX PROGRAMMING By Anatoli B. Juditsky and Arkadi S. Nemirovski1 Universit´eGrenoble I and Georgia Institute of Technology The problem we concentrate on is as follows: given (1) a convex compact set X in Rn, an affine mapping x 7→ A(x), a parametric family {pµ(·)} of probability densities and (2) N i.i.d. observations of the random variable ω, distributed with the density pA(x)(·) for some (unknown) x ∈ X, estimate the value gT x of a given linear form at x. For several families {pµ(·)} with no additional assumptions on X and A, we develop computationally efficient estimation routines which are minimax optimal, within an absolute constant factor. We then apply these routines to recovering x itself in the Euclidean norm. 1. Introduction. The problem we are interested in is essentially as fol- lows: suppose that we are given a convex compact set X in Rn, an affine mapping x A(x) and a parametric family pµ( ) of probability densities. Suppose that7→N i.i.d. observations of the random{ variable· } ω, distributed with the density p ( ) for some (unknown) x X, are available. Our objective A(x) · ∈ is to estimate the value gT x of a given linear form at x. In nonparametric statistics, there exists an immense literature on various versions of this problem (see, e.g., [10, 11, 12, 13, 15, 17, 18, 21, 22, 23, 24, 25, 26, 27, 28] and the references therein). -

Statistical Analysis in JASP
Copyright © 2018 by Mark A Goss-Sampson. All rights reserved. This book or any portion thereof may not be reproduced or used in any manner whatsoever without the express written permission of the author except for the purposes of research, education or private study. CONTENTS PREFACE .................................................................................................................................................. 1 USING THE JASP INTERFACE .................................................................................................................... 2 DESCRIPTIVE STATISTICS ......................................................................................................................... 8 EXPLORING DATA INTEGRITY ................................................................................................................ 15 ONE SAMPLE T-TEST ............................................................................................................................. 22 BINOMIAL TEST ..................................................................................................................................... 25 MULTINOMIAL TEST .............................................................................................................................. 28 CHI-SQUARE ‘GOODNESS-OF-FIT’ TEST............................................................................................. 30 MULTINOMIAL AND Χ2 ‘GOODNESS-OF-FIT’ TEST. .......................................................................... -

Friedman Test
community project encouraging academics to share statistics support resources All stcp resources are released under a Creative Commons licence stcp-marquier-FriedmanR The following resources are associated: The R dataset ‘Video.csv’ and R script ‘Friedman.R’ Friedman test in R (Non-parametric equivalent to repeated measures ANOVA) Dependent variable: Continuous (scale) but not normally distributed or ordinal Independent variable: Categorical (Time/ Condition) Common Applications: Used when several measurements of the same dependent variable are taken at different time points or under different conditions for each subject and the assumptions of repeated measures ANOVA have not been met. It can also be used to compare ranked outcomes. Data: The dataset ‘Video’ contains some results from a study comparing videos made to aid understanding of a particular medical condition. Participants watched three videos (A, B, C) and one product demonstration (D) and were asked several Likert style questions about each. These were summed to give an overall score for each e.g. TotalAGen below is the total score of the ordinal questions for video A. Friedman ranks each participants responses The Friedman test ranks each person’s score from lowest to highest (as if participants had been asked to rank the methods from least favourite to favourite) and bases the test on the sum of ranks for each column. For example, person 1 gave C the lowest Total score of 13 and A the highest so SPSS would rank these as 1 and 4 respectively. As the raw data is ranked to carry out the test, the Friedman test can also be used for data which is already ranked e.g. -

Compare Multiple Related Samples
Compare Multiple Related Samples The command compares multiple related samples using the Friedman test (nonparametric alternative to the one-way ANOVA with repeated measures) and calculates the Kendall's coefficient of concordance (also known as Kendall’s W). Kendall's W makes no assumptions about the underlying probability distribution and allows to handle any number of outcomes, unlike the standard Pearson correlation coefficient. Friedman test is similar to the Kruskal-Wallis one-way analysis of variance with the difference that Friedman test is an alternative to the repeated measures ANOVA with balanced design. How To For unstacked data (each column is a sample): o Run the STATISTICS->NONPARAMETRIC STATISTICS -> COMPARE MULTIPLE RELATED SAMPLES [FRIEDMAN ANOVA, CONCORDANCE] command. o Select variables to compare. For stacked data (with a group variable): o Run the STATISTICS->NONPARAMETRIC STATISTICS -> COMPARE MULTIPLE RELATED SAMPLES (WITH GROUP VARIABLE) command. o Select a variable with observations (VARIABLE) and a text or numeric variable with the group names (GROUPS). RESULTS The report includes Friedman ANOVA and Kendall’s W test results. THE FRIEDMAN ANOVA tests the null hypothesis that the samples are from identical populations. If the p-value is less than the selected 훼 level the null-hypothesis is rejected. If there are no ties, Friedman test statistic Ft is defined as: 푘 12 퐹 = [ ∑ 푅2] − 3푛(푘 + 1) 푡 푛푘(푘 + 1) 푖 푖=1 where n is the number of rows, or subjects; k is the number of columns or conditions, and Ri is the sum of the ranks of ith column. If ranking results in any ties, the Friedman test statistic Ft is defined as: 푅2 푛(푘 − 1) [∑푘 푖 − 퐶 ] 푖=1 푛 퐹 퐹푡 = 2 ∑ 푟푖푗 − 퐶퐹 where n is the number rows, or subjects, k is the number of columns, and Ri is the sum of the ranks from column, or condition I; CF is the ties correction (Corder et al., 2009). -

Repeated-Measures ANOVA & Friedman Test Using STATCAL
Repeated-Measures ANOVA & Friedman Test Using STATCAL (R) & SPSS Prana Ugiana Gio Download STATCAL in www.statcal.com i CONTENT 1.1 Example of Case 1.2 Explanation of Some Book About Repeated-Measures ANOVA 1.3 Repeated-Measures ANOVA & Friedman Test 1.4 Normality Assumption and Assumption of Equality of Variances (Sphericity) 1.5 Descriptive Statistics Based On SPSS dan STATCAL (R) 1.6 Normality Assumption Test Using Kolmogorov-Smirnov Test Based on SPSS & STATCAL (R) 1.7 Assumption Test of Equality of Variances Using Mauchly Test Based on SPSS & STATCAL (R) 1.8 Repeated-Measures ANOVA Based on SPSS & STATCAL (R) 1.9 Multiple Comparison Test Using Boferroni Test Based on SPSS & STATCAL (R) 1.10 Friedman Test Based on SPSS & STATCAL (R) 1.11 Multiple Comparison Test Using Wilcoxon Test Based on SPSS & STATCAL (R) ii 1.1 Example of Case For example given data of weight of 11 persons before and after consuming medicine of diet for one week, two weeks, three weeks and four week (Table 1.1.1). Tabel 1.1.1 Data of Weight of 11 Persons Weight Name Before One Week Two Weeks Three Weeks Four Weeks A 89.43 85.54 80.45 78.65 75.45 B 85.33 82.34 79.43 76.55 71.35 C 90.86 87.54 85.45 80.54 76.53 D 91.53 87.43 83.43 80.44 77.64 E 90.43 84.45 81.34 78.64 75.43 F 90.52 86.54 85.47 81.44 78.64 G 87.44 83.34 80.54 78.43 77.43 H 89.53 86.45 84.54 81.35 78.43 I 91.34 88.78 85.47 82.43 78.76 J 88.64 84.36 80.66 78.65 77.43 K 89.51 85.68 82.68 79.71 76.5 Average 89.51 85.68 82.68 79.71 76.69 Based on Table 1.1.1: The person whose name is A has initial weight 89,43, after consuming medicine of diet for one week 85,54, two weeks 80,45, three weeks 78,65 and four weeks 75,45. -
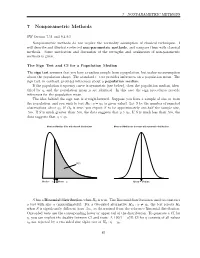
7 Nonparametric Methods
7 NONPARAMETRIC METHODS 7 Nonparametric Methods SW Section 7.11 and 9.4-9.5 Nonparametric methods do not require the normality assumption of classical techniques. I will describe and illustrate selected non-parametric methods, and compare them with classical methods. Some motivation and discussion of the strengths and weaknesses of non-parametric methods is given. The Sign Test and CI for a Population Median The sign test assumes that you have a random sample from a population, but makes no assumption about the population shape. The standard t−test provides inferences on a population mean. The sign test, in contrast, provides inferences about a population median. If the population frequency curve is symmetric (see below), then the population median, iden- tified by η, and the population mean µ are identical. In this case the sign procedures provide inferences for the population mean. The idea behind the sign test is straightforward. Suppose you have a sample of size m from the population, and you wish to test H0 : η = η0 (a given value). Let S be the number of sampled observations above η0. If H0 is true, you expect S to be approximately one-half the sample size, .5m. If S is much greater than .5m, the data suggests that η > η0. If S is much less than .5m, the data suggests that η < η0. Mean and Median differ with skewed distributions Mean and Median are the same with symmetric distributions 50% Median = η Mean = µ Mean = Median S has a Binomial distribution when H0 is true. The Binomial distribution is used to construct a test with size α (approximately). -

Chapter 4. Data Analysis
4. DATA ANALYSIS Data analysis begins in the monitoring program group of observations selected from the target design phase. Those responsible for monitoring population. In the case of water quality should identify the goals and objectives for monitoring, descriptive statistics of samples are monitoring and the methods to be used for used almost invariably to formulate conclusions or analyzing the collected data. Monitoring statistical inferences regarding populations objectives should be specific statements of (MacDonald et al., 1991; Mendenhall, 1971; measurable results to be achieved within a stated Remington and Schork, 1970; Sokal and Rohlf, time period (Ponce, 1980b). Chapter 2 provides 1981). A point estimate is a single number an overview of commonly encountered monitoring representing the descriptive statistic that is objectives. Once goals and objectives have been computed from the sample or group of clearly established, data analysis approaches can observations (Freund, 1973). For example, the be explored. mean total suspended solids concentration during baseflow is 35 mg/L. Point estimates such as the Typical data analysis procedures usually begin mean (as in this example), median, mode, or with screening and graphical methods, followed geometric mean from a sample describe the central by evaluating statistical assumptions, computing tendency or location of the sample. The standard summary statistics, and comparing groups of data. deviation and interquartile range could likewise be The analyst should take care in addressing the used as point estimates of spread or variability. issues identified in the information expectations report (Section 2.2). By selecting and applying The use of point estimates is warranted in some suitable methods, the data analyst responsible for cases, but in nonpoint source analyses point evaluating the data can prevent the “data estimates of central tendency should be coupled rich)information poor syndrome” (Ward 1996; with an interval estimate because of the large Ward et al., 1986). -

Basic Statistical Analysis Aov(), Anova(), Lm(), Glm(): Linear And
basic statistical analysis R reference card, by Jonathan Baron aov(), anova(), lm(), glm(): linear and non- Parentheses are for functions, brackets are for indi- linear models, anova cating the position of items in a vector or matrix. t.test(): t test (Here, items with numbers like x1 are user-supplied variables.) prop.test(), binom.test(): sign test chisq.test(x1): chi-square test on matrix x1 Miscellaneous fisher.test(): Fisher exact test q(): quit cor(a): show correlations <-: assign cor.test(a,b): test correlation INSTALL package1: install package1 friedman.test(): Friedman test m1[,2]: column 2 of matrix m1 m1[,2:5] or m1[,c(2,3,4,5)]: columns 2–5 some statistics in mva package m1$a1: variable a1 in data frame m1 prcomp(): principal components NA: missing data kmeans(): kmeans cluster analysis is.na: true if data missing factanal(): factor analysis library(mva): load (e.g.) the mva package cancor(): canonical correlation Help Graphics help(command1): get help with command1 (NOTE: plot(), barplot(), boxplot(), stem(), hist(): USE THIS FOR MORE DETAIL THAN THIS basic plots CARD CAN PROVIDE.) matplot(): matrix plot help.start(): start browser help pairs(matrix): scatterplots help(package=mva): help with (e.g.) package mva coplot(): conditional plot apropos("topic1"): commands relevant to topic1 stripchart(): strip chart example(command1): examples of command1 qqplot(): quantile-quantile plot Input and output qqnorm(), qqline(): fit normal distribution source("file1"): run the commands in file1. read.table("file1"): read in data from file1 -

1 Exercise 8: an Introduction to Descriptive and Nonparametric
1 Exercise 8: An Introduction to Descriptive and Nonparametric Statistics Elizabeth M. Jakob and Marta J. Hersek Goals of this lab 1. To understand why statistics are used 2. To become familiar with descriptive statistics 1. To become familiar with nonparametric statistical tests, how to conduct them, and how to choose among them 2. To apply this knowledge to sample research questions Background If we are very fortunate, our experiments yield perfect data: all the animals in one treatment group behave one way, and all the animals in another treatment group behave another way. Usually our results are not so clear. In addition, even if we do get results that seem definitive, there is always a possibility that they are a result of chance. Statistics enable us to objectively evaluate our results. Descriptive statistics are useful for exploring, summarizing, and presenting data. Inferential statistics are used for interpreting data and drawing conclusions about our hypotheses. Descriptive statistics include the mean (average of all of the observations; see Table 8.1), mode (most frequent data class), and median (middle value in an ordered set of data). The variance, standard deviation, and standard error are measures of deviation from the mean (see Table 8.1). These statistics can be used to explore your data before going on to inferential statistics, when appropriate. In hypothesis testing, a variety of statistical tests can be used to determine if the data best fit our null hypothesis (a statement of no difference) or an alternative hypothesis. More specifically, we attempt to reject one of these hypotheses. -

Nonparametric Analogues of Analysis of Variance
*\ NONPARAMETRIC ANALOGUES OF ANALYSIS OF VARIANCE by °\\4 RONALD K. LOHRDING B. A., Southwestern College, 1963 A MASTER'S REPORT submitted in partial fulfillment of the requirements for the degree MASTER OF SCIENCE Department of Statistics and Statistical Laboratory KANSAS STATE UNIVERSITY Manhattan, Kansas 1966 Approved by: ' UKC )x 0-i Major PrProfessor L IVoJL CONTENTS 1. INTRODUCTION 1 2. COMPLETELY RANDOMIZED DESIGN 4 2. 1. The Kruskal-Wallis One-way Analysis of Variance by Ranks 4 2. 2. Mosteller's k-Sample Slippage Test 7 2.3. Conover's k-Sample Slippage Test 8 2.4. A k-Sample Kolmogorov-Smirnov Test 11 2 2. 5. The Contingency x Test 13 3. RANDOMIZED COMPLETE BLOCK 14 3. 1. Cochran Q Test 15 3.2. Friedman's Two-way Analysis of Variance by Ranks . 16 4. PROCEDURES WHICH GENERALIZE TO SEVERAL DESIGNS . 17 4.1. Mood's Median Test 18 4.2. Wilson's Median Test 21 4.3. The Randomized Rank-Sum Test 27 4.4. Rank Test for Paired-Comparison Experiments ... 30 5. BALANCED INCOMPLETE BLOCK 33 5. 1. Bradley's Rank Analysis of Incomplete Block Designs 33 5.2. Durbin's Rank Analysis of Incomplete Block Designs 40 6. A 2x2 FACTORIAL EXPERIMENT 44 7. PARTIALLY BALANCED INCOMPLETE BLOCK 47 OTHER RELATED TESTS 50 ACKNOWLEDGEMENTS 53 REFERENCES 54 NONPARAMETRIC ANALOGUES OF ANALYSIS OF VARIANCE 1. Introduction J. V. Bradley (I960) proposes that the history of statistics can be divided into four major stages. The first or one parameter stage was when statistics were merely thought of as averages or in some instances ratios.