(Ade1) with Hysteresis Behavior and Quorum-Quenching Activity
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

MIAMI UNIVERSITY the Graduate School Certificate for Approving The
MIAMI UNIVERSITY The Graduate School Certificate for Approving the Dissertation We hereby approve the Dissertation of Matthew F. Rouhier Candidate for the Degree: Doctor of Philosophy ___________________________________________________________ Director (Dr. Ann Hagerman) ___________________________________________________________ Reader (Dr.Chris Makaroff) ___________________________________________________________ Reader (Dr.Gary Lorigan) __________________________________________________________ Reader (Dr. Richard Taylor) _________________________________________________________ Graduate School Representative (Dr. Paul James) ! ABSTRACT CHARACTERIZATION OF YDR036C FROM Saccharomyces cerevisiae by Matthew F. Rouhier Beta-hydroxyisobutyryl-CoA (HIBYL-CoA) hydrolases are found ubiquitously in eukaryotes where they function in the catabolism of valine. A homologous enzyme (YDR036C) is also present in yeast where valine catabolism is distinctly different and does not require a HIBYL-CoA hydrolase. Like the other eukaryotic hydrolases, the yeast hydrolase is a member of the crotonase super-family which catalyzes various reactions using CoA thioester substrates. Crotonases typically function in fatty acid degradation which is peroxisomal in yeast while YDR036C is found strictly in mitochondria. Like other HIBYL-CoA hydrolases the yeast enzyme demonstrated activity toward beta-hydroxyacyl-CoAs, but unlike the other HIBYL-CoA hydrolases the yeast activity is greatest with beta-hydroxypropionyl-CoA (3-HP-CoA). Further characterization of the active site has determined that the preference of the hydrolases for 3-HP- CoA or HIBYL-CoA is dependent upon the residue at position 177. The glutamate at position 121 is responsible for the coordination with the beta-hydroxyl group and replacement with valine renders the enzyme non-specific for a hydroxyl group. Another unique feature to yeast hydrolases is the presence of a C-terminal tail of 80 amino acids, which when removed renders the enzyme inactive, suggesting that it may play a role in substrate binding. -

Yeast Genome Gazetteer P35-65
gazetteer Metabolism 35 tRNA modification mitochondrial transport amino-acid metabolism other tRNA-transcription activities vesicular transport (Golgi network, etc.) nitrogen and sulphur metabolism mRNA synthesis peroxisomal transport nucleotide metabolism mRNA processing (splicing) vacuolar transport phosphate metabolism mRNA processing (5’-end, 3’-end processing extracellular transport carbohydrate metabolism and mRNA degradation) cellular import lipid, fatty-acid and sterol metabolism other mRNA-transcription activities other intracellular-transport activities biosynthesis of vitamins, cofactors and RNA transport prosthetic groups other transcription activities Cellular organization and biogenesis 54 ionic homeostasis organization and biogenesis of cell wall and Protein synthesis 48 plasma membrane Energy 40 ribosomal proteins organization and biogenesis of glycolysis translation (initiation,elongation and cytoskeleton gluconeogenesis termination) organization and biogenesis of endoplasmic pentose-phosphate pathway translational control reticulum and Golgi tricarboxylic-acid pathway tRNA synthetases organization and biogenesis of chromosome respiration other protein-synthesis activities structure fermentation mitochondrial organization and biogenesis metabolism of energy reserves (glycogen Protein destination 49 peroxisomal organization and biogenesis and trehalose) protein folding and stabilization endosomal organization and biogenesis other energy-generation activities protein targeting, sorting and translocation vacuolar and lysosomal -

Thioesterase Superfamily Member 1 Undergoes Stimulus-Coupled Conformational Reorganization to Regulate Metabolism in Mice
ARTICLE https://doi.org/10.1038/s41467-021-23595-x OPEN Thioesterase superfamily member 1 undergoes stimulus-coupled conformational reorganization to regulate metabolism in mice Yue Li 1,2, Norihiro Imai3, Hayley T. Nicholls 3, Blaine R. Roberts 4, Samaksh Goyal 1,2, Tibor I. Krisko 3, Lay-Hong Ang 1,2, Matthew C. Tillman4, Anne M. Roberts4, Mahnoor Baqai 1,2, Eric A. Ortlund 4, ✉ ✉ David E. Cohen 3 & Susan J. Hagen 1,2 1234567890():,; In brown adipose tissue, thermogenesis is suppressed by thioesterase superfamily member 1 (Them1), a long chain fatty acyl-CoA thioesterase. Them1 is highly upregulated by cold ambient temperature, where it reduces fatty acid availability and limits thermogenesis. Here, we show that Them1 regulates metabolism by undergoing conformational changes in response to β-adrenergic stimulation that alter Them1 intracellular distribution. Them1 forms metabolically active puncta near lipid droplets and mitochondria. Upon stimulation, Them1 is phosphorylated at the N-terminus, inhibiting puncta formation and activity and resulting in a diffuse intracellular localization. We show by correlative light and electron microscopy that Them1 puncta are biomolecular condensates that are inhibited by phosphorylation. Thus, Them1 forms intracellular biomolecular condensates that limit fatty acid oxidation and sup- press thermogenesis. During a period of energy demand, the condensates are disrupted by phosphorylation to allow for maximal thermogenesis. The stimulus-coupled reorganization of Them1 provides fine-tuning of thermogenesis and energy expenditure. 1 Division of General Surgery, Department of Surgery, Beth Israel Deaconess Medical Center, Boston, MA, USA. 2 Department of Surgery, Harvard Medical School, Boston, MA, USA. 3 Division of Gastroenterology and Hepatology, Department of Medicine, Weill Cornell Medical College, New York, NY, USA. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Inflammatory Stimuli Induce Acyl-Coa Thioesterase 7 and Remodeling of Phospholipids Containing Unsaturated Long (C20)-Acyl Chains in Macrophages
Supplemental Material can be found at: http://www.jlr.org/content/suppl/2017/04/17/jlr.M076489.DC1 .html Inflammatory stimuli induce acyl-CoA thioesterase 7 and remodeling of phospholipids containing unsaturated long (C20)-acyl chains in macrophages Valerie Z. Wall,*,† Shelley Barnhart,* Farah Kramer,* Jenny E. Kanter,* Anuradha Vivekanandan-Giri,§ Subramaniam Pennathur,§ Chiara Bolego,** Jessica M. Ellis,§§,*** Miguel A. Gijón,††† Michael J. Wolfgang,*** and Karin E. Bornfeldt1,*,† Department of Medicine,* Division of Metabolism, Endocrinology and Nutrition, and Department of Pathology,† UW Medicine Diabetes Institute, University of Washington, Seattle, WA; Department of Internal Medicine,§ University of Michigan, Ann Arbor, MI; Department of Pharmaceutical and Pharmacological Sciences,** University of Padova, Padova, Italy; Department of Nutrition Science,§§ Purdue University, West Lafayette, IN; Department of Biological Chemistry,*** Johns Hopkins University School of Medicine, Baltimore, MD; and Department of Pharmacology,††† University of Downloaded from Colorado Denver, Aurora, CO Abstract Acyl-CoA thioesterase 7 (ACOT7) is an intracel- containing unsaturated long (C20)-acyl chains in macro- lular enzyme that converts acyl-CoAs to FFAs. ACOT7 is in- phages, and, although ACOT7 has preferential thioesterase duced by lipopolysaccharide (LPS); thus, we investigated activity toward these lipid species, loss of ACOT7 has no ma- www.jlr.org downstream effects of LPS-induced induction of ACOT7 jor detrimental effect on macrophage inflammatory pheno- and its role in inflammatory settings in myeloid cells. Enzy- types.—Wall, V. Z., S. Barnhart, F. Kramer, J. E. Kanter, matic thioesterase activity assays in WT and ACOT7-deficient A. Vivekanandan-Giri, S. Pennathur, C. Bolego, J. M. Ellis, macrophage lysates indicated that endogenous ACOT7 con- M. -

The Metabolic Serine Hydrolases and Their Functions in Mammalian Physiology and Disease Jonathan Z
REVIEW pubs.acs.org/CR The Metabolic Serine Hydrolases and Their Functions in Mammalian Physiology and Disease Jonathan Z. Long* and Benjamin F. Cravatt* The Skaggs Institute for Chemical Biology and Department of Chemical Physiology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, California 92037, United States CONTENTS 2.4. Other Phospholipases 6034 1. Introduction 6023 2.4.1. LIPG (Endothelial Lipase) 6034 2. Small-Molecule Hydrolases 6023 2.4.2. PLA1A (Phosphatidylserine-Specific 2.1. Intracellular Neutral Lipases 6023 PLA1) 6035 2.1.1. LIPE (Hormone-Sensitive Lipase) 6024 2.4.3. LIPH and LIPI (Phosphatidic Acid-Specific 2.1.2. PNPLA2 (Adipose Triglyceride Lipase) 6024 PLA1R and β) 6035 2.1.3. MGLL (Monoacylglycerol Lipase) 6025 2.4.4. PLB1 (Phospholipase B) 6035 2.1.4. DAGLA and DAGLB (Diacylglycerol Lipase 2.4.5. DDHD1 and DDHD2 (DDHD Domain R and β) 6026 Containing 1 and 2) 6035 2.1.5. CES3 (Carboxylesterase 3) 6026 2.4.6. ABHD4 (Alpha/Beta Hydrolase Domain 2.1.6. AADACL1 (Arylacetamide Deacetylase-like 1) 6026 Containing 4) 6036 2.1.7. ABHD6 (Alpha/Beta Hydrolase Domain 2.5. Small-Molecule Amidases 6036 Containing 6) 6027 2.5.1. FAAH and FAAH2 (Fatty Acid Amide 2.1.8. ABHD12 (Alpha/Beta Hydrolase Domain Hydrolase and FAAH2) 6036 Containing 12) 6027 2.5.2. AFMID (Arylformamidase) 6037 2.2. Extracellular Neutral Lipases 6027 2.6. Acyl-CoA Hydrolases 6037 2.2.1. PNLIP (Pancreatic Lipase) 6028 2.6.1. FASN (Fatty Acid Synthase) 6037 2.2.2. PNLIPRP1 and PNLIPR2 (Pancreatic 2.6.2. -

(10) Patent No.: US 8119385 B2
US008119385B2 (12) United States Patent (10) Patent No.: US 8,119,385 B2 Mathur et al. (45) Date of Patent: Feb. 21, 2012 (54) NUCLEICACIDS AND PROTEINS AND (52) U.S. Cl. ........................................ 435/212:530/350 METHODS FOR MAKING AND USING THEMI (58) Field of Classification Search ........................ None (75) Inventors: Eric J. Mathur, San Diego, CA (US); See application file for complete search history. Cathy Chang, San Diego, CA (US) (56) References Cited (73) Assignee: BP Corporation North America Inc., Houston, TX (US) OTHER PUBLICATIONS c Mount, Bioinformatics, Cold Spring Harbor Press, Cold Spring Har (*) Notice: Subject to any disclaimer, the term of this bor New York, 2001, pp. 382-393.* patent is extended or adjusted under 35 Spencer et al., “Whole-Genome Sequence Variation among Multiple U.S.C. 154(b) by 689 days. Isolates of Pseudomonas aeruginosa” J. Bacteriol. (2003) 185: 1316 1325. (21) Appl. No.: 11/817,403 Database Sequence GenBank Accession No. BZ569932 Dec. 17. 1-1. 2002. (22) PCT Fled: Mar. 3, 2006 Omiecinski et al., “Epoxide Hydrolase-Polymorphism and role in (86). PCT No.: PCT/US2OO6/OOT642 toxicology” Toxicol. Lett. (2000) 1.12: 365-370. S371 (c)(1), * cited by examiner (2), (4) Date: May 7, 2008 Primary Examiner — James Martinell (87) PCT Pub. No.: WO2006/096527 (74) Attorney, Agent, or Firm — Kalim S. Fuzail PCT Pub. Date: Sep. 14, 2006 (57) ABSTRACT (65) Prior Publication Data The invention provides polypeptides, including enzymes, structural proteins and binding proteins, polynucleotides US 201O/OO11456A1 Jan. 14, 2010 encoding these polypeptides, and methods of making and using these polynucleotides and polypeptides. -

The Microbiota-Produced N-Formyl Peptide Fmlf Promotes Obesity-Induced Glucose
Page 1 of 230 Diabetes Title: The microbiota-produced N-formyl peptide fMLF promotes obesity-induced glucose intolerance Joshua Wollam1, Matthew Riopel1, Yong-Jiang Xu1,2, Andrew M. F. Johnson1, Jachelle M. Ofrecio1, Wei Ying1, Dalila El Ouarrat1, Luisa S. Chan3, Andrew W. Han3, Nadir A. Mahmood3, Caitlin N. Ryan3, Yun Sok Lee1, Jeramie D. Watrous1,2, Mahendra D. Chordia4, Dongfeng Pan4, Mohit Jain1,2, Jerrold M. Olefsky1 * Affiliations: 1 Division of Endocrinology & Metabolism, Department of Medicine, University of California, San Diego, La Jolla, California, USA. 2 Department of Pharmacology, University of California, San Diego, La Jolla, California, USA. 3 Second Genome, Inc., South San Francisco, California, USA. 4 Department of Radiology and Medical Imaging, University of Virginia, Charlottesville, VA, USA. * Correspondence to: 858-534-2230, [email protected] Word Count: 4749 Figures: 6 Supplemental Figures: 11 Supplemental Tables: 5 1 Diabetes Publish Ahead of Print, published online April 22, 2019 Diabetes Page 2 of 230 ABSTRACT The composition of the gastrointestinal (GI) microbiota and associated metabolites changes dramatically with diet and the development of obesity. Although many correlations have been described, specific mechanistic links between these changes and glucose homeostasis remain to be defined. Here we show that blood and intestinal levels of the microbiota-produced N-formyl peptide, formyl-methionyl-leucyl-phenylalanine (fMLF), are elevated in high fat diet (HFD)- induced obese mice. Genetic or pharmacological inhibition of the N-formyl peptide receptor Fpr1 leads to increased insulin levels and improved glucose tolerance, dependent upon glucagon- like peptide-1 (GLP-1). Obese Fpr1-knockout (Fpr1-KO) mice also display an altered microbiome, exemplifying the dynamic relationship between host metabolism and microbiota. -

Cholesterol Esterase from Porcine Pancreas
Cholesterol Esterase from porcine pancreas Catalog Number C9464 Storage Temperature –20 °C CAS RN 9026-00-0 Substrates: EC 3.1.1.13 cholesteryl esters17 triacylglycerol2 Synonyms: Bile salt activated lipase, sterol esterase, 4-nitrophenyl butyrate7 triolein9 carboxyl ester lipase, steryl-ester acylhydrolase 2-(diethylamino)-4H-3,1-benzoxazin-4-one18 2-(diethylamino)-4H-thieno[2,3-d]oxazin-4-one18 Product Description 7 Excess cholesterol is stored intracellularly as KM: 4-nitrophenyl butyrate cholesterol esters. Cholesterol esterase (CE) is a 0.37 mM (with taurocholate) reversible enzyme that can hydrolyze or synthesize 0.73 mM (without taurocholate) fatty acid esters of cholesterol and other sterols. Hydrolysis of water insoluble long chain fatty acid Activators: esters requires bile salt activation. Hydrolysis of water ethanol9 methanol9 soluble esters of short chain fatty acids and n-butanol9 sodium taurocholate10 lysophospholipids does not require activation by bile sodium cholate11 salts.1 Cholesterol esterase catalyzes the following reaction: Inhibitors: bisphenol A methacrylate12 Hg2+,15 Cholesterol Cholesterol Cholesterol diisopropylfluorophosphate13 enolase14 esters esterase + Fatty acid sodium fluoride15 phosphatidic acid16 phosphatidylcholine16 phosphatidylserine16 While found primarily in the pancreas and pancreatic 4-nitrophenyl-N-substituted carbamates11 fluid, it occurs in other tissues as well. In the bovine adrenal cortex, this reaction is one of the rate limiting Ki: steps in steroidogenesis, involving the release of 4-nitrophenyl-N-allyl -

Computational Modeling of Solvent Effects on Protein-Ligand
Commun. Comput. Phys. Vol. 13, No. 1, pp. 31-60 doi: 10.4208/cicp.130911.121011s January 2013 Computational Modeling of Solvent Effects on Protein-Ligand Interactions Using Fully Polarizable Continuum Model and Rational Drug Design Fang Zheng and Chang-Guo Zhan∗ Department of Pharmaceutical Sciences, College of Pharmacy, University of Kentucky, 789 South Limestone Street, Lexington, Kentucky 40536, USA. Received 13 September 2011; Accepted (in revised version) 12 October 2011 Available online 12 June 2012 Abstract. This is a brief review of the computational modeling of protein-ligand in- teractions using a recently developed fully polarizable continuum model (FPCM) and rational drug design. Computational modeling has become a powerful tool in under- standing detailed protein-ligand interactions at molecular level and in rational drug design. To study the binding of a protein with multiple molecular species of a ligand, one must accurately determine both the relative free energies of all of the molecular species in solution and the corresponding microscopic binding free energies for all of the molecular species binding with the protein. In this paper, we aim to provide a brief overview of the recent development in computational modeling of the solvent ef- fects on the detailed protein-ligand interactions involving multiple molecular species of a ligand related to rational drug design. In particular, we first briefly discuss the main challenges in computational modeling of the detailed protein-ligand interactions involving the multiple molecular species and then focus on the FPCM model and its applications. The FPCM method allows accurate determination of the solvent effects in the first-principles quantum mechanism (QM) calculations on molecules in solu- tion. -
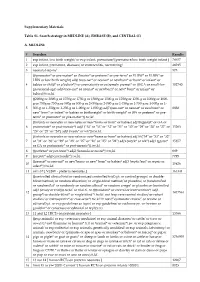
EMBASE (B), and CENTRAL (C) A. MEDLINE # Searches
Supplementary Materials Table S1. Search strategy in MEDLINE (A), EMBASE (B), and CENTRAL (C) A. MEDLINE # Searches Results 1 exp infant, low birth weight/ or exp infant, premature/ [premature/low birth weight infant ] 78657 2 exp infant, premature, diseases/ or enterocolitis, necrotizing/ 46015 3 neonatal sepsis/ 575 (((prematur* or pre-matur* or i?matur* or preterm* or pre-term* or VLBW* or ELBW* or LBW or low birth weight) adj6 (neo-nat* or neonat* or newborn* or born* or infant* or 4 babies or child* or p?ediatr*)) or prematurity or extremely premat* or ((SGA or small-for- 102740 gestational-age) adj6 (neo-nat* or neonat* or newborn* or new* born* or infant* or babies))).tw,kf. ((2000g or 2000-g or 1750g or 1750-g or 1500g or 1500-g or 1250g or 1250-g or 1000g or 1000- g or 750g or 750-g or 500g or 500-g or 2-000g or 2-000-g or 1-750g or 1-750-g or 1-500g or 1- 5 500-g or 1-250g or 1-250-g or 1-000g or 1-000-g) adj7 (neo-nat* or neonat* or newborn* or 8838 new* born* or infant* or babies or birthweight* or birth weight* or BW or preterm* or pre- term* or prematur* or pre-matur*)).tw,kf. ((infants or neonates or neo-nates or new*borns or born* or babies) adj18 (gestat* or GA or 6 postmenstr* or post-menstr*) adj3 ("34" or "33" or "32" or "31" or "30" or "29" or "28" or "27" or 15263 "26" or "25" or "24") adj3 (week* or wk*)).tw,kf. -

September, 2008
NIDA - Director's Report - September, 2008 NIDA Home > Publications > Director's Reports Director's Report to the National Advisory Council on Drug Abuse - September, 2008 Index Research Findings Cross-Divisional Research Basic Neurosciences Research Basic Behavioral Research Behavioral and Brain Development Research Clinical Neuroscience Research Epidemiology and Etiology Research Prevention Research Research on Behavioral and Combined Treatments for Drug Abuse Research on Pharmacotherapies for Drug Abuse Research on Medical Consequences of Drug Abuse and Co-Occurring Infections (HIV/AIDS, HCV) Services Research Clinical Trials Network Research International Research Intramural Research Program Activities Extramural Policy and Review Activities Congressional Affairs International Activities Meetings and Conferences Media and Education Activities Planned Meetings Publications Staff Highlights Grantee Honors https://archives.drugabuse.gov/DirReports/DirRep908/Default.html[11/17/16, 11:11:47 PM] NIDA - Director's Report - September, 2008 Archive Home | Accessibility | Privacy | FOIA (NIH) | Current NIDA Home Page The National Institute on Drug Abuse (NIDA) is part of the National Institutes of Health (NIH) , a component of the U.S. Department of Health and Human Services. Questions? _ See our Contact Information. https://archives.drugabuse.gov/DirReports/DirRep908/Default.html[11/17/16, 11:11:47 PM] NIDA - Director's Report - September, 2008 NIDA Home > Publications > Director's Reports > September, 2008 Index Director's Report to the National