Functional Study of the Role Played by Nucleolar Proteins in the Control of Neural Progenitor Homeostasis Using Zebrafish As a Model Alessandro Brombin
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Preclinical Research 2020 Sections
Sackler Faculty of Medicine Preclinical Research 2020 Sections Cancer and Molecular Therapies 8 Dental Health and Medicine 52 Diabetes, Metabolic and Endocrine Diseases 62 Genomics & Personalized Medicine 78 Hearing, Language & Speech Sciences and Disorders 98 Infectious Diseases 117 Inflammatory and Autoimmune Diseases 137 Medical Education and Ethics 147 Nervous System and Brain Disorders 154 Nursing, Occupational and Physical Therapy 200 Public Health 235 Reproduction, Development and Evolution 263 Stem Cells and Regenerative Medicine 280 Cover images (from bottom left, clockwise): Image 1: Human embryonic stem cell derived cardiomyocytes stained with fluorescent antibodies. The cardiac marker alpha-actinin (green), calcium channel modulator, Ahnak1 (red) – Shimrit Oz, Nathan Dascal. Image 2: Islet of Langerhans containing insulin-producing beta-cells (green) and glucagon- producing alpha-cells (red) – Daria Baer, Limor Landsman. Image 3: β-catenin in C. elegans vulva – Michal Caspi, Limor Broday, Rina Rosin-Arbesfeld. Image 4: Stereocilia of a sensory outer hair cell from a mouse inner ear – Shaked Shivatzki, Karen Avraham. Image 5: Electron scanning micrograph of middle ear ossicles from a mouse ear stained with pseudo colors – Shaked Shivatski, Karen Avraham. Image 6: Resistin-like molecule alpha (red), eosinophil major basic protein (green) and DAPI (blue) staining of asthmatic mice – Danielle Karo-Atar, Ariel Munitz. © All rights reserved Editor: Prof. Karen Avraham Graphic design: Michal Semo Kovetz, TAU Graphic Design Studio February 2020 Sackler Faculty of Medicine Research 2020 2 The Sackler Faculty of Medicine The Sackler Faculty of Medicine is Israel’s largest diabetes, neurodegenerative diseases, infectious medical research and training complex. The Sackler diseases and genetic diseases, including but not Faculty of Medicine of Tel Aviv University (TAU) was imited to Alzheimer’s disease, Parkinson’s disease founded in 1964 following the generous contributions and HIV/AIDS. -

Introduction to the Cell Cell History Cell Structures and Functions
Introduction to the cell cell history cell structures and functions CK-12 Foundation December 16, 2009 CK-12 Foundation is a non-profit organization with a mission to reduce the cost of textbook materials for the K-12 market both in the U.S. and worldwide. Using an open-content, web-based collaborative model termed the “FlexBook,” CK-12 intends to pioneer the generation and distribution of high quality educational content that will serve both as core text as well as provide an adaptive environment for learning. Copyright ©2009 CK-12 Foundation This work is licensed under the Creative Commons Attribution-Share Alike 3.0 United States License. To view a copy of this license, visit http://creativecommons.org/licenses/by-sa/3.0/us/ or send a letter to Creative Commons, 171 Second Street, Suite 300, San Francisco, California, 94105, USA. Contents 1 Cell structure and function dec 16 5 1.1 Lesson 3.1: Introduction to Cells .................................. 5 3 www.ck12.org www.ck12.org 4 Chapter 1 Cell structure and function dec 16 1.1 Lesson 3.1: Introduction to Cells Lesson Objectives • Identify the scientists that first observed cells. • Outline the importance of microscopes in the discovery of cells. • Summarize what the cell theory proposes. • Identify the limitations on cell size. • Identify the four parts common to all cells. • Compare prokaryotic and eukaryotic cells. Introduction Knowing the make up of cells and how cells work is necessary to all of the biological sciences. Learning about the similarities and differences between cell types is particularly important to the fields of cell biology and molecular biology. -
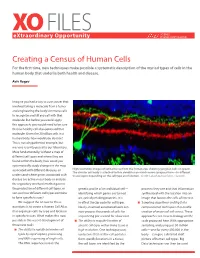
Creating a Census of Human Cells
XO FILES SCIENCE eXtraordinary Opportunity PHILANTHROPY ALLIANCE Creating a Census of Human Cells For the first time, new techniques make possible a systematic description of the myriad types of cells in the human body that underlie both health and disease. Aviv Regev Imagine you had a way to cure cancer that involved taking a molecule from a tumor and engineering the body’s immune cells to recognize and kill any cell with that molecule. But before you could apply this approach, you would need to be sure that no healthy cell also expressed that molecule. Given the 20 trillion cells in a human body, how would you do that? This is not a hypothetical example, but was one recently posed to our laboratory. More fundamentally, without a map of different cell types and where they are found within the body, how could you systematically study changes in the map associated with different diseases, or High resolution images of retinal tissue from the human eye, showing ganglion cells (in green). The circular cell body is attached to thin dendrites on which nerve synapses form—in different understand where genes associated with tissue layers depending on the cell type and function. Credit: Lab of Joshua Sanes, Harvard. disease are active in our body or analyze the regulatory mechanisms that govern the production of different cell types, or genetic profile of an individual cell— proteins they use and that information sort out how different cell types combine identifying which genes are turned synthesized with the location into an to form specific tissues? on, actively making proteins. -

The Ribosomal Protein Rpl22 Controls Ribosome Composition by Directly Repressing Expression of Its Own Paralog, Rpl22l1
The Ribosomal Protein Rpl22 Controls Ribosome Composition by Directly Repressing Expression of Its Own Paralog, Rpl22l1 Monique N. O’Leary1,2, Katherine H. Schreiber1,2, Yong Zhang3, Anne-Ce´cile E. Duc3, Shuyun Rao3,J. Scott Hale4¤, Emmeline C. Academia2, Shreya R. Shah1, John F. Morton5, Carly A. Holstein6, Dan B. Martin6, Matt Kaeberlein7, Warren C. Ladiges5, Pamela J. Fink4, Vivian L. MacKay1, David L. Wiest3, Brian K. Kennedy1,2* 1 Department of Biochemistry, University of Washington, Seattle, Washington, United States of America, 2 Buck Institute for Research on Aging, Novato, California, United States of America, 3 Blood Cell Development and Cancer Keystone, Immune Cell Development and Host Defense Program, Fox Chase Cancer Center, Philadelphia, Pennsylvania, United States of America, 4 Department of Immunology, University of Washington, Seattle, Washington, United States of America, 5 Department of Comparative Medicine, University of Washington, Seattle, Washington, United States of America, 6 Institute for Systems Biology, Seattle, Washington, United States of America, 7 Department of Pathology, University of Washington, Seattle, Washington, United States of America Abstract Most yeast ribosomal protein genes are duplicated and their characterization has led to hypotheses regarding the existence of specialized ribosomes with different subunit composition or specifically-tailored functions. In yeast, ribosomal protein genes are generally duplicated and evidence has emerged that paralogs might have specific roles. Unlike yeast, most mammalian ribosomal proteins are thought to be encoded by a single gene copy, raising the possibility that heterogenous populations of ribosomes are unique to yeast. Here, we examine the roles of the mammalian Rpl22, finding that Rpl222/2 mice have only subtle phenotypes with no significant translation defects. -

Assessment of Hematopoietic Failure Due to Rpl11 Deficiency in a Zebrafish Model of Diamond-Blackfan Anemia by Deep Sequencing
Zhang et al. BMC Genomics 2013, 14:896 http://www.biomedcentral.com/1471-2164/14/896 RESEARCH ARTICLE Open Access Assessment of hematopoietic failure due to Rpl11 deficiency in a zebrafish model of Diamond- Blackfan anemia by deep sequencing Zhaojun Zhang1†, Haibo Jia2†, Qian Zhang1, Yang Wan3, Yang Zhou2, Qiong Jia2, Wanguang Zhang4, Weiping Yuan3, Tao Cheng3, Xiaofan Zhu3* and Xiangdong Fang1* Abstract Background: Diamond–Blackfan anemia is a rare congenital red blood cell dysplasia that develops soon after birth. RPL11 mutations account for approximately 4.8% of human DBA cases with defective hematopoietic phenotypes. However, the mechanisms by which RPL11 regulates hematopoiesis in DBA remain elusive. In this study, we analyzed the transcriptome using deep sequencing data from an Rpl11-deficient zebrafish model to identify Rpl11-mediated hematopoietic failure and investigate the underlying mechanisms. Results: We characterized hematological defects in Rpl11-deficient zebrafish embryos by identifying affected hematological genes, hematopoiesis-associated pathways, and regulatory networks. We found that hemoglobin biosynthetic and hematological defects in Rpl11-deficient zebrafish were related to dysregulation of iron metabolism-related genes, including tfa, tfr1b, alas2 and slc25a37, which are involved in heme and hemoglobin biosynthesis. In addition, we found reduced expression of the hematopoietic stem cells (HSC) marker cmyb and HSC transcription factors tal1 and hoxb4a in Rpl11-deficient zebrafish embryos, indicating that the hematopoietic defects may be related to impaired HSC formation, differentiation, and proliferation. However, Rpl11 deficiency did not affect the development of other blood cell lineages such as granulocytes and myelocytes. Conclusion: We identified hematopoietic failure of Rpl11-deficient zebrafish embryos using transcriptome deep sequencing and elucidated potential underlying mechanisms. -

Overexpressed WDR3 Induces the Activation of Hippo Pathway by Interacting with GATA4 in Pancreatic Cancer
Overexpressed WDR3 Induces the Activation of Hippo Pathway by Interacting with GATA4 in Pancreatic Cancer Wenjie Su Sichuan Provincial People's Hospital: Sichuan Academy of Medical Sciences and Sichuan People's Hospital Shikai Zhu Sichuan Provincial People's Hospital: Sichuan Academy of Medical Sciences and Sichuan People's Hospital Kai Chen Sichuan Provincial People's Hospital: Sichuan Academy of Medical Sciences and Sichuan People's Hospital Hongji Yang Sichuan Provincial People's Hospital: Sichuan Academy of Medical Sciences and Sichuan People's Hospital Mingwu Tian Sichuan Provincial People's Hospital: Sichuan Academy of Medical Sciences and Sichuan People's Hospital Qiang Fu Massachusetts General Hospital Ganggang Shi University of British Columbia School of Human Kinetics: The University of British Columbia School of Kinesiology Shijian Feng University of British Columbia School of Human Kinetics: The University of British Columbia School of Kinesiology Dianyun Ren Wuhan Union Hospital Xin Jin Wuhan Union Hospital Chong Yang ( [email protected] ) Sichuan Provincial People's Hospital: Sichuan Academy of Medical Sciences and Sichuan People's Hospital Page 1/34 Research Keywords: Pancreatic Cancer, WDR3, GATA4, YAP1, Hippo Signaling Pathway Posted Date: November 13th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-104564/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Version of Record: A version of this preprint was published on March 1st, 2021. See the published version at https://doi.org/10.1186/s13046-021-01879-w. Page 2/34 Abstract Background: WD repeat domain 3 (WDR3) is involved in a variety of cellular processes including gene regulation, cell cycle progression, signal transduction and apoptosis. -

Identification of the Key Genes and Pathways in Prostate Cancer
ONCOLOGY LETTERS 16: 6663-6669, 2018 Identification of the key genes and pathways in prostate cancer SHUTONG FAN1*, ZUMU LIANG1*, ZHIQIN GAO1, ZHIWEI PAN2, SHAOJIE HAN3, XIAOYING LIU1, CHUNLING ZHAO1, WEIWEI YANG1, ZHIFANG PAN1 and WEIGUO FENG1 1College of Bioscience and Technology, Weifang Medical University, Weifang, Shandong 261053; 2Department of Internal Medicine, Laizhou Development Zone Hospital, Yantai, Shandong 261400; 3Animal Epidemic Prevention and Epidemic Control Center, Changle County Bureau of Animal Health and Production, Weifang, Shandong 262400, P.R. China Received March 5, 2018; Accepted September 17, 2018 DOI: 10.3892/ol.2018.9491 Abstract. Prostate cancer (PCa) is one of the most common Introduction malignancies in men globally. The aim of the present study was to identify the key genes and pathways involved in the Prostate cancer (PCa) is one of the most common malignancies occurrence of PCa. Gene expression profile (GSE55945) in men globally and the second leading cause of cancer was downloaded from Gene Expression Omnibus, and associated mortality in developed countries (1,2). Like other the differentially expressed genes (DEGs) were identified. cancers, PCa is considered to be a disease which caused by Subsequently, Gene ontology analysis, KEGG pathway age, diet and gene aberrations (3). Accumulating evidences analysis and protein-protein interaction (PPI) analysis of have demonstrated that a series of genes and pathways involved DEGs were performed. Finally, the identified key genes were in the occurrence, progression and metastasis of PCa (4). At confirmed by immunohistochemistry. The GO analysis results present, the underlying mechanism of PCa occurrence is still showed that the DEGs were mainly participated in cell cycle, unclear, which limits the diagnosis and therapy. -

Variation in Protein Coding Genes Identifies Information
bioRxiv preprint doi: https://doi.org/10.1101/679456; this version posted June 21, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Animal complexity and information flow 1 1 2 3 4 5 Variation in protein coding genes identifies information flow as a contributor to 6 animal complexity 7 8 Jack Dean, Daniela Lopes Cardoso and Colin Sharpe* 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 Institute of Biological and Biomedical Sciences 25 School of Biological Science 26 University of Portsmouth, 27 Portsmouth, UK 28 PO16 7YH 29 30 * Author for correspondence 31 [email protected] 32 33 Orcid numbers: 34 DLC: 0000-0003-2683-1745 35 CS: 0000-0002-5022-0840 36 37 38 39 40 41 42 43 44 45 46 47 48 49 Abstract bioRxiv preprint doi: https://doi.org/10.1101/679456; this version posted June 21, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Animal complexity and information flow 2 1 Across the metazoans there is a trend towards greater organismal complexity. How 2 complexity is generated, however, is uncertain. Since C.elegans and humans have 3 approximately the same number of genes, the explanation will depend on how genes are 4 used, rather than their absolute number. -

Robustness and Timing of Cellular Differentiation Through Population-Based Symmetry Breaking
AR TI CLE Robustness AND TIMING OF CELLULAR DIFFERENTIATION THROUGH population-based SYMMETRY BREAKING Angel StanoeV1 | Christian Schröter1 | Aneta KOSESKA1∗ 1Department OF Systemic Cell Biology, Max Planck Institute OF Molecular PhYSIOLOGY, Although COORDINATED BEHAVIOR OF MULTICELLULAR ORGANISMS Dortmund, GermanY RELIES ON INTERCELLULAR communication, THE DYNAMICAL mech- Correspondence ANISM THAT UNDERLIES SYMMETRY BREAKING IN HOMOGENEOUS Email: populations, THEREBY GIVING RISE TO HETEROGENEOUS CELL TYPES ∗[email protected] REMAINS UNCLEAR. In CONTRAST TO THE PREVALENT cell-intrinsic VIEW OF DIFFERENTIATION WHERE MULTISTABILITY ON THE SINGLE CELL LEVEL IS A NECESSARY pre-requisite, WE PROPOSE THAT het- EROGENEOUS CELLULAR IDENTITIES EMERGE AND ARE MAINTAINED COOPERATIVELY ON A POPULATION LEVel. Identifying A GENERIC SYMMETRY BREAKING mechanism, WE DEMONSTRATE THAT ROBUST PROPORTIONS BETWEEN DIFFERENTIATED CELL TYPES ARE INTRINSIC FEATURES OF THIS INHOMOGENEOUS STEADY STATE solution. Crit- ICAL ORGANIZATION IN THE VICINITY OF THE SYMMETRY BREAKING BIFURCATION ON THE OTHER hand, SUGGESTS THAT TIMING OF cel- LULAR DIFFERENTIATION EMERGES FOR GROWING POPULATIONS IN A self-organized MANNER. Setting A CLEAR DISTINCTION BETWEEN pre-eXISTING ASYMMETRIES AND SYMMETRY BREAKING EVents, WE THUS DEMONSTRATE THAT EMERGENT SYMMETRY breaking, IN CONJUNCTION WITH ORGANIZATION NEAR CRITICALITY NECESSARILY LEADS TO ROBUSTNESS AND ACCURACY DURING DEVelopment. KEYWORDS SYMMETRY breaking; differentiation; INHOMOGENEOUS STEADY STATE Abbreviations: -

Download Report 2010-12
RESEARCH REPORt 2010—2012 MAX-PLANCK-INSTITUT FÜR WISSENSCHAFTSGESCHICHTE Max Planck Institute for the History of Science Cover: Aurora borealis paintings by William Crowder, National Geographic (1947). The International Geophysical Year (1957–8) transformed research on the aurora, one of nature’s most elusive and intensely beautiful phenomena. Aurorae became the center of interest for the big science of powerful rockets, complex satellites and large group efforts to understand the magnetic and charged particle environment of the earth. The auroral visoplot displayed here provided guidance for recording observations in a standardized form, translating the sublime aesthetics of pictorial depictions of aurorae into the mechanical aesthetics of numbers and symbols. Most of the portait photographs were taken by Skúli Sigurdsson RESEARCH REPORT 2010—2012 MAX-PLANCK-INSTITUT FÜR WISSENSCHAFTSGESCHICHTE Max Planck Institute for the History of Science Introduction The Max Planck Institute for the History of Science (MPIWG) is made up of three Departments, each administered by a Director, and several Independent Research Groups, each led for five years by an outstanding junior scholar. Since its foundation in 1994 the MPIWG has investigated fundamental questions of the history of knowl- edge from the Neolithic to the present. The focus has been on the history of the natu- ral sciences, but recent projects have also integrated the history of technology and the history of the human sciences into a more panoramic view of the history of knowl- edge. Of central interest is the emergence of basic categories of scientific thinking and practice as well as their transformation over time: examples include experiment, ob- servation, normalcy, space, evidence, biodiversity or force. -
![Arxiv:1910.00443V3 [Eess.IV] 29 Jul 2020 Synchronize Acquired Data Sets, Such That Corresponding Developmental Stages Are Compared to One Another [3, 6]](https://docslib.b-cdn.net/cover/9873/arxiv-1910-00443v3-eess-iv-29-jul-2020-synchronize-acquired-data-sets-such-that-corresponding-developmental-stages-are-compared-to-one-another-3-6-709873.webp)
Arxiv:1910.00443V3 [Eess.IV] 29 Jul 2020 Synchronize Acquired Data Sets, Such That Corresponding Developmental Stages Are Compared to One Another [3, 6]
Towards Automatic Embryo Staging in 3D+t Microscopy Images using Convolutional Neural Networks and PointNets? Manuel Traub1;2[0000−0003−0897−1701] and Johannes Stegmaier1[0000−0003−4072−3759] 1 Institute of Imaging and Computer Vision, RWTH Aachen University, Germany 2 Institute for Automation and Applied Informatics, Karlsruhe Institute of Technology, Germany [email protected] Abstract. Automatic analyses and comparisons of different stages of embryonic development largely depend on a highly accurate spatiotem- poral alignment of the investigated data sets. In this contribution, we assess multiple approaches for automatic staging of developing embryos that were imaged with time-resolved 3D light-sheet microscopy. The methods comprise image-based convolutional neural networks as well as an approach based on the PointNet architecture that directly operates on 3D point clouds of detected cell nuclei centroids. The experiments with four wild-type zebrafish embryos render both approaches suitable for au- tomatic staging with average deviations of 21 − 34 minutes. Moreover, a proof-of-concept evaluation based on simulated 3D+t point cloud data sets shows that average deviations of less than 7 minutes are possible. Keywords: Convolutional Neural Networks · PointNet · Regression · Transfer Learning · Developmental Biology · Simulating Embryogenesis. 1 Introduction Embryonic development is characterized by a multitude of synchronized events, cell shape changes and large-scale tissue rearrangements that are crucial steps in the successful formation of a new organism [10]. To be able to compare these developmental events among different wild-type individuals, different mutants or upon exposure to certain chemicals, it is highly important to temporally arXiv:1910.00443v3 [eess.IV] 29 Jul 2020 synchronize acquired data sets, such that corresponding developmental stages are compared to one another [3, 6]. -

Deep Multiomics Profiling of Brain Tumors Identifies Signaling Networks
ARTICLE https://doi.org/10.1038/s41467-019-11661-4 OPEN Deep multiomics profiling of brain tumors identifies signaling networks downstream of cancer driver genes Hong Wang 1,2,3, Alexander K. Diaz3,4, Timothy I. Shaw2,5, Yuxin Li1,2,4, Mingming Niu1,4, Ji-Hoon Cho2, Barbara S. Paugh4, Yang Zhang6, Jeffrey Sifford1,4, Bing Bai1,4,10, Zhiping Wu1,4, Haiyan Tan2, Suiping Zhou2, Laura D. Hover4, Heather S. Tillman 7, Abbas Shirinifard8, Suresh Thiagarajan9, Andras Sablauer 8, Vishwajeeth Pagala2, Anthony A. High2, Xusheng Wang 2, Chunliang Li 6, Suzanne J. Baker4 & Junmin Peng 1,2,4 1234567890():,; High throughput omics approaches provide an unprecedented opportunity for dissecting molecular mechanisms in cancer biology. Here we present deep profiling of whole proteome, phosphoproteome and transcriptome in two high-grade glioma (HGG) mouse models driven by mutated RTK oncogenes, PDGFRA and NTRK1, analyzing 13,860 proteins and 30,431 phosphosites by mass spectrometry. Systems biology approaches identify numerous master regulators, including 41 kinases and 23 transcription factors. Pathway activity computation and mouse survival indicate the NTRK1 mutation induces a higher activation of AKT down- stream targets including MYC and JUN, drives a positive feedback loop to up-regulate multiple other RTKs, and confers higher oncogenic potency than the PDGFRA mutation. A mini-gRNA library CRISPR-Cas9 validation screening shows 56% of tested master regulators are important for the viability of NTRK-driven HGG cells, including TFs (Myc and Jun) and metabolic kinases (AMPKa1 and AMPKa2), confirming the validity of the multiomics inte- grative approaches, and providing novel tumor vulnerabilities.