Investigating Hypoxic Tumor Physiology Through Gene Expression Patterns
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

(12) Patent Application Publication (10) Pub. No.: US 2009/0111743 A1 Takahashi (43) Pub
US 200901 11743A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2009/0111743 A1 Takahashi (43) Pub. Date: Apr. 30, 2009 (54) CYSTEINE-BRANCHED HEPARIN-BINDING Related U.S. Application Data GROWTH FACTOR ANALOGS (60) Provisional application No. 60/656,713, filed on Feb. 25, 2005. (75) Inventor: Kazuyuki Takahashi, O O Germantown, MD (US) Publication Classification (51) Int. Cl. Correspondence Address: A638/08C07K 700 30.82006.O1 201 THIRD STREET, N.W., SUITE 1340 A638/16 (2006.01) ALBUQUEROUE, NM 87102 (US) A638/10 (2006.01) A638/00 (2006.01) (73) Assignee: BioSurface Engineering (52) U.S. Cl. ........... 514/12:530/330; 530/329; 530/328; Technologies, Inc., Rockville, MD 530/327: 530/326; 530/325; 530/324; 514/17; (US) 514/16; 514/15: 514/14: 514/13 (57) ABSTRACT (21) Appl. No.: 11/362,137 The present invention provides a heparin binding growth factor analog of any of formula I-VIII and methods and uses (22) Filed: Feb. 23, 2006 thereof. US 2009/011 1743 A1 Apr. 30, 2009 CYSTENE-BRANCHED HEPARN-BINDING 0007 HBGFs useful in prevention or therapy of a wide GROWTH FACTOR ANALOGS range of diseases and disorders may be purified from natural sources or produced by recombinant DNA methods, however, CROSS-REFERENCE TO RELATED Such preparations are expensive and generally difficult to APPLICATIONS prepare. 0008. Some efforts have been made to generate heparin 0001. This application claims priority to and the benefit of binding growth factor analogs. For example, natural PDGF the filing of U.S. Provisional Patent Application Ser. -

Angiocrine Endothelium: from Physiology to Cancer Jennifer Pasquier1,2*, Pegah Ghiabi2, Lotf Chouchane3,4,5, Kais Razzouk1, Shahin Rafi3 and Arash Rafi1,2,3
Pasquier et al. J Transl Med (2020) 18:52 https://doi.org/10.1186/s12967-020-02244-9 Journal of Translational Medicine REVIEW Open Access Angiocrine endothelium: from physiology to cancer Jennifer Pasquier1,2*, Pegah Ghiabi2, Lotf Chouchane3,4,5, Kais Razzouk1, Shahin Rafi3 and Arash Rafi1,2,3 Abstract The concept of cancer as a cell-autonomous disease has been challenged by the wealth of knowledge gathered in the past decades on the importance of tumor microenvironment (TM) in cancer progression and metastasis. The sig- nifcance of endothelial cells (ECs) in this scenario was initially attributed to their role in vasculogenesis and angiogen- esis that is critical for tumor initiation and growth. Nevertheless, the identifcation of endothelial-derived angiocrine factors illustrated an alternative non-angiogenic function of ECs contributing to both physiological and pathological tissue development. Gene expression profling studies have demonstrated distinctive expression patterns in tumor- associated endothelial cells that imply a bilateral crosstalk between tumor and its endothelium. Recently, some of the molecular determinants of this reciprocal interaction have been identifed which are considered as potential targets for developing novel anti-angiocrine therapeutic strategies. Keywords: Angiocrine, Endothelium, Cancer, Cancer microenvironment, Angiogenesis Introduction of blood vessels in initiation of tumor growth and stated Metastatic disease accounts for about 90% of patient that in the absence of such angiogenesis, tumors can- mortality. Te difculty in controlling and eradicating not expand their mass or display a metastatic phenotype metastasis might be related to the heterotypic interaction [7]. Based on this theory, many investigators assumed of tumor and its microenvironment [1]. -

Type of the Paper (Article
Table S1. Gene expression of pro-angiogenic factors in tumor lymph nodes of Ibtk+/+Eµ-myc and Ibtk+/-Eµ-myc mice. Fold p- Symbol Gene change value 0,007 Akt1 Thymoma viral proto-oncogene 1 1,8967 061 0,929 Ang Angiogenin, ribonuclease, RNase A family, 5 1,1159 481 0,000 Angpt1 Angiopoietin 1 4,3916 117 0,461 Angpt2 Angiopoietin 2 0,7478 625 0,258 Anpep Alanyl (membrane) aminopeptidase 1,1015 737 0,000 Bai1 Brain-specific angiogenesis inhibitor 1 4,0927 202 0,001 Ccl11 Chemokine (C-C motif) ligand 11 3,1381 149 0,000 Ccl2 Chemokine (C-C motif) ligand 2 2,8407 298 0,000 Cdh5 Cadherin 5 2,5849 744 0,000 Col18a1 Collagen, type XVIII, alpha 1 3,8568 388 0,003 Col4a3 Collagen, type IV, alpha 3 2,9031 327 0,000 Csf3 Colony stimulating factor 3 (granulocyte) 4,3332 258 0,693 Ctgf Connective tissue growth factor 1,0195 88 0,000 Cxcl1 Chemokine (C-X-C motif) ligand 1 2,67 21 0,067 Cxcl2 Chemokine (C-X-C motif) ligand 2 0,7507 631 0,000 Cxcl5 Chemokine (C-X-C motif) ligand 5 3,921 328 0,000 Edn1 Endothelin 1 3,9931 042 0,001 Efna1 Ephrin A1 1,6449 601 0,002 Efnb2 Ephrin B2 2,8858 042 0,000 Egf Epidermal growth factor 1,726 51 0,000 Eng Endoglin 0,2309 467 0,000 Epas1 Endothelial PAS domain protein 1 2,8421 764 0,000 Ephb4 Eph receptor B4 3,6334 035 V-erb-b2 erythroblastic leukemia viral oncogene homolog 2, 0,000 Erbb2 3,9377 neuro/glioblastoma derived oncogene homolog (avian) 024 0,000 F2 Coagulation factor II 3,8295 239 1 0,000 F3 Coagulation factor III 4,4195 293 0,002 Fgf1 Fibroblast growth factor 1 2,8198 748 0,000 Fgf2 Fibroblast growth factor -
Targeting Fibroblast Growth Factor Pathways in Endometrial Cancer
Curr Probl Cancer 41 (2017) 37–47 Contents lists available at ScienceDirect Curr Probl Cancer journal homepage: www.elsevier.com/locate/cpcancer Targeting fibroblast growth factor pathways in endometrial cancer Boris Winterhoff, MDa, Gottfried E. Konecny, MDb,* article info abstract Novel treatments that improve outcomes for patients with Keywords: recurrent or metastatic endometrial cancer (EC) remain an Endometrial cancer unmet need. Aberrant signaling by fibroblast growth factors Fibroblast growth factor (FGFs) and FGF receptors (FGFRs) has been implicated in Angiogenesis several human cancers. Activating mutations in FGFR2 have been found in up to 16% of ECs, suggesting an opportunity for targeted therapy. This review summarizes the role of the FGF pathway in angiogenesis and EC, and provides an overview of FGFR-targeted therapies under clinical develop- ment for the treatment of EC. & 2017 The Authors. Published by Elsevier Inc. This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/). Introduction Endometrial cancer (EC) is the most common gynecologic cancer, with 54,870 new cases and 10,170 related deaths estimated for the United States in 2015.1 Localized disease is often curable with surgery and the 5-year relative survival rate for patients diagnosed with early-stage EC is high.1-3 However, for patients who present with metastatic disease or experience a recurrence, prognosis is poor.1-3 Response of metastatic EC to standard therapies (eg, adjuvant radiotherapy, brachytherapy, or chemotherapy) is limited and overall survival for most patients with recurrent or metastatic disease is r1year.2-4 Thus, novel treatment options for this disease remain an unmet need. -
Endothelial Cells and VEGF in Vascular Development Leigh Coultas1, Kallayanee Chawengsaksophak1 & Janet Rossant1
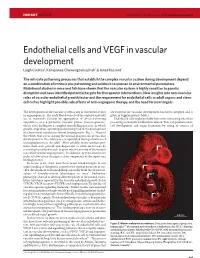
Insight Rossant p7-15 6/12/05 10:23 AM Page 9377 INSIGHT REVIEW NATURE|Vol 438|15 December 2005|doi:10.1038/nature04479 Endothelial cells and VEGF in vascular development Leigh Coultas1, Kallayanee Chawengsaksophak1 & Janet Rossant1 The intricate patterning processes that establish the complex vascular system during development depend on a combination of intrinsic pre-patterning and extrinsic responses to environmental parameters. Mutational studies in mice and fish have shown that the vascular system is highly sensitive to genetic disruption and have identified potential targets for therapeutic interventions. New insights into non-vascular roles of vascular endothelial growth factor and the requirement for endothelial cells in adult organs and stem- cell niches highlight possible side effects of anti-angiogenic therapy and the need for new targets. The development of the vascular system is one of the earliest events are essential for vascular development has been compiled and is in organogenesis. The early blood vessels of the embryo and yolk given in Supplementary Table 1. sac in mammals develop by aggregation of de-novo-forming Endothelial cells might probably have more interesting roles than angioblasts into a primitive vascular plexus (vasculogenesis), just acting as channels for blood circulation. They can promote stem- which then undergoes a complex remodelling process, in which cell development and organ formation by acting as sources of growth, migration, sprouting and pruning lead to the development of a functional circulatory system (angiogenesis; Fig. 1). Many of the events that occur during the normal progression of vascular development in the embryo are recapitulated during situations of neoangiogenesis in the adult1. -

Vascular Endothelial Growth Factor (VEGF) Signaling in Tumor Progression Robert Roskoski Jr
Critical Reviews in Oncology/Hematology 62 (2007) 179–213 Vascular endothelial growth factor (VEGF) signaling in tumor progression Robert Roskoski Jr. ∗ Blue Ridge Institute for Medical Research, 3754 Brevard Road, Suite 116A, Box 19, Horse Shoe, NC 28742, USA Accepted 29 January 2007 Contents 1. Vasculogenesis and angiogenesis....................................................................................... 180 1.1. Definitions .................................................................................................... 180 1.2. Physiological and non-physiological angiogenesis ................................................................. 181 1.3. Activators and inhibitors of angiogenesis ......................................................................... 181 1.4. Sprouting and non-sprouting angiogenesis ........................................................................ 181 1.5. Tumor vessel morphology....................................................................................... 183 2. The vascular endothelial growth factor (VEGF) family ................................................................... 183 3. Properties and expression of the VEGF family .......................................................................... 183 3.1. VEGF-A ...................................................................................................... 183 3.2. VEGF-B ...................................................................................................... 185 3.3. VEGF-C ..................................................................................................... -
Figure S1. Reverse Transcription‑Quantitative PCR Analysis of ETV5 Mrna Expression Levels in Parental and ETV5 Stable Transfectants
Figure S1. Reverse transcription‑quantitative PCR analysis of ETV5 mRNA expression levels in parental and ETV5 stable transfectants. (A) Hec1a and Hec1a‑ETV5 EC cell lines; (B) Ishikawa and Ishikawa‑ETV5 EC cell lines. **P<0.005, unpaired Student's t‑test. EC, endometrial cancer; ETV5, ETS variant transcription factor 5. Figure S2. Survival analysis of sample clusters 1‑4. Kaplan Meier graphs for (A) recurrence‑free and (B) overall survival. Survival curves were constructed using the Kaplan‑Meier method, and differences between sample cluster curves were analyzed by log‑rank test. Figure S3. ROC analysis of hub genes. For each gene, ROC curve (left) and mRNA expression levels (right) in control (n=35) and tumor (n=545) samples from The Cancer Genome Atlas Uterine Corpus Endometrioid Cancer cohort are shown. mRNA levels are expressed as Log2(x+1), where ‘x’ is the RSEM normalized expression value. ROC, receiver operating characteristic. Table SI. Clinicopathological characteristics of the GSE17025 dataset. Characteristic n % Atrophic endometrium 12 (postmenopausal) (Control group) Tumor stage I 91 100 Histology Endometrioid adenocarcinoma 79 86.81 Papillary serous 12 13.19 Histological grade Grade 1 30 32.97 Grade 2 36 39.56 Grade 3 25 27.47 Myometrial invasiona Superficial (<50%) 67 74.44 Deep (>50%) 23 25.56 aMyometrial invasion information was available for 90 of 91 tumor samples. Table SII. Clinicopathological characteristics of The Cancer Genome Atlas Uterine Corpus Endometrioid Cancer dataset. Characteristic n % Solid tissue normal 16 Tumor samples Stagea I 226 68.278 II 19 5.740 III 70 21.148 IV 16 4.834 Histology Endometrioid 271 81.381 Mixed 10 3.003 Serous 52 15.616 Histological grade Grade 1 78 23.423 Grade 2 91 27.327 Grade 3 164 49.249 Molecular subtypeb POLE 17 7.328 MSI 65 28.017 CN Low 90 38.793 CN High 60 25.862 CN, copy number; MSI, microsatellite instability; POLE, DNA polymerase ε. -

Assessment of Plasma Cytokine Profile Changes in Bevacizumab
Biochemistry and Molecular Biology Assessment of Plasma Cytokine Profile Changes in Bevacizumab-Treated Retinopathy of Prematurity Infants Lingkun Kong,1 Ann B. Demny,2 Ahmar Sajjad,1 Amit R. Bhatt,1 and Sridevi Devaraj3 1Department of Ophthalmology, Baylor College of Medicine, Houston, Texas, United States 2Department of Pediatrics, Baylor College of Medicine, Houston, Texas, United States 3Department of Clinical Pathology, Baylor College of Medicine, Houston, Texas, United States Correspondence: Lingkun Kong, De- PURPOSE. We compared changes of plasma angiogenesis cytokine profiles in infants who were partment of Ophthalmology, Baylor treated with intravitreal injection of bevacizumab (IVB) for type 1 retinopathy of prematurity College of Medicine, 6701 Fannin (ROP) with age-matched preterm non-ROP infants. Street,Suite610.25,Houston,TX 77030, USA; METHODS. Thirteen infants with type 1 ROP and 13 age-matched preterm non-ROP infants [email protected]. were included. Blood samples were collected prior to treatment (time 0) and 6 weeks after Submitted: October 28, 2015 the treatment (time 42). Plasma levels of nine cytokines from the angiogenesis growth factor Accepted: February 21, 2016 panel and seven soluble cytokine receptors were measured using a magnetic multiplex assay. Citation: Kong L, Demny AB, Sajjad A, RESULTS. Plasma cytokine profiles changed from time 0 to time 42 in both groups. In Bhatt AR, Devaraj S. Assessment of bevacizumab-treated ROP infants, the following plasma angiogenesis growth factor and plasma cytokine profile changes in soluble cytokine receptor levels decreased significantly: soluble VEGF-A (sVEGF-A; P ¼ bevacizumab-treated retinopathy of 0.0001), sVEGF-D (P ¼ 0.04), angiopoietin-2 (Ang-2; P ¼ 0.002), sVEGF receptor 1 (R1) and prematurity infants. -

Plgf) in the Antitumor Immune Response and Cancer Progression
International Journal of Molecular Sciences Review Multifaceted Role of the Placental Growth Factor (PlGF) in the Antitumor Immune Response and Cancer Progression Loredana Albonici *, Maria Gabriella Giganti, Andrea Modesti, Vittorio Manzari and Roberto Bei Department of Clinical Sciences and Translational Medicine, University of Rome “Tor Vergata”, Via Montpellier 1, 00133 Rome, Italy; [email protected] (M.G.G.); [email protected] (A.M.); [email protected] (V.M.); [email protected] (R.B.) * Correspondence: [email protected] Received: 8 May 2019; Accepted: 14 June 2019; Published: 18 June 2019 Abstract: The sharing of molecules function that affects both tumor growth and neoangiogenesis with cells of the immune system creates a mutual interplay that impairs the host’s immune response against tumor progression. Increasing evidence shows that tumors are able to create an immunosuppressive microenvironment by recruiting specific immune cells. Moreover, molecules produced by tumor and inflammatory cells in the tumor microenvironment create an immunosuppressive milieu able to inhibit the development of an efficient immune response against cancer cells and thus fostering tumor growth and progression. In addition, the immunoediting could select cancer cells that are less immunogenic or more resistant to lysis. In this review, we summarize recent findings regarding the immunomodulatory effects and cancer progression of the angiogenic growth factor namely placental growth factor (PlGF) and address the biological complex effects of this cytokine. Different pathways of the innate and adaptive immune response in which, directly or indirectly, PlGF is involved in promoting tumor immune escape and metastasis will be described. PlGF is important for building up vascular structures and functions. -

Placental Growth Factor As a Marker of Fetal Growth Restriction Caused by Placental Dysfunction
Placenta 42 (2016) 1e8 Contents lists available at ScienceDirect Placenta journal homepage: www.elsevier.com/locate/placenta Placental growth factor as a marker of fetal growth restriction caused by placental dysfunction * Samantha J. Benton a, , Lesley M. McCowan b, Alexander E.P. Heazell c, David Grynspan d, Jennifer A. Hutcheon a, Christof Senger e, Orlaith Burke f, Yuen Chan b, Jane E. Harding g, Julien Yockell-Lelievre h, Yuxiang Hu a, Lucy C. Chappell i, Melanie J. Griffin i, Andrew H. Shennan i, Laura A. Magee a, 1, Andree Gruslin j, 2, Peter von Dadelszen a, 1 a Department of Obstetrics and Gynaecology, Faculty of Medicine, University of British Columbia, 4500 Oak Street, Vancouver, British Columbia, V6H 3N1, Canada b Department of Obstetrics and Gynaecology, Faculty of Medical and Health Sciences, University of Auckland, Private Bag 92019, Auckland, 1142, New Zealand c Maternal and Fetal Health Research Centre, University of Manchester, 5th Floor, St. Mary's Hospital, Oxford Road, Manchester, M13 9WL, United Kingdom d Department of Pathology, Children's Hospital of Eastern Ontario, 401 Smyth Road, Ottawa, Ontario, K1H 8L1, Canada e Department of Pathology, Children's and Women's Health Centre of British Columbia, 4500 Oak Street, Vancouver, British Columbia, V6H 2N9, Canada f Nuffield Department of Population Health, Institute of Health Sciences, University of Oxford, Old Road, Oxford, OX3 7LF, United Kingdom g Liggins Institute, University of Auckland, Private Bag 92019, Victoria Street West, Auckland, 1142, New Zealand h Sprott -
List of Predicted Circfam120a Target Mrnas Gene in Pathway Species Gene Name
List of predicted circFAM120A target mRNAs Gene in pathway Species Gene name HRAS Homo sapiens HRas proto-oncogene, GTPase (HRAS) ADCY1 Homo sapiens Adenylate cyclase 1 (ADCY1) ADCY2 Homo sapiens Adenylate cyclase 2 (ADCY2) ADCY7 Homo sapiens Adenylate cyclase 7 (ADCY7) PTGS2 Homo sapiens Prostaglandin-endoperoxide synthase 2 (PTGS2) PGF Homo sapiens Placental growth factor (PGF) ADCY5 Homo sapiens Adenylate cyclase 5 (ADCY5) STAT5A Homo sapiens Signal transducer and activator of transcription 5A (STAT5A) ADCY6 Homo sapiens Adenylate cyclase 6 (ADCY6) STAT5B Homo sapiens Signal transducer and activator of transcription 5B (STAT5B) LPAR3 Homo sapiens Lysophosphatidic acid receptor 3 (LPAR3) LPAR2 Homo sapiens Lysophosphatidic acid receptor 2 (LPAR2) CTNNB1 Homo sapiens Catenin beta 1 (CTNNB1) CUL2 Homo sapiens Cullin 2 (CUL2) RARA Homo sapiens Retinoic acid receptor alpha (RARA) GNG2 Homo sapiens G protein subunit gamma 2 (GNG2) RARB Homo sapiens Retinoic acid receptor beta (RARB) GNG3 Homo sapiens G protein subunit gamma 3 (GNG3) FAS Homo sapiens Fas cell surface death receptor (FAS) GNG4 Homo sapiens G protein subunit gamma 4 (GNG4) GNG7 Homo sapiens G protein subunit gamma 7 (GNG7) PIK3CG Homo sapiens Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma (PIK3CG) WNT10B Homo sapiens Wnt family member 10B (WNT10B) BCR Homo sapiens BCR, RhoGEF and GTPase activating protein (BCR) BRAF Homo sapiens B-Raf proto-oncogene, serine/threonine kinase (BRAF) RXRB Homo sapiens Retinoid X receptor beta (RXRB) ROCK2 Homo sapiens Rho -

Prediction of Stillbirth from Placental Growth Factor at 11--13 Weeks
Ultrasound Obstet Gynecol 2016; 48: 618–623 Published online in Wiley Online Library (wileyonlinelibrary.com). DOI: 10.1002/uog.17288 Prediction of stillbirth from placental growth factor at 11–13 weeks R. AKOLEKAR*†, M. MACHUCA*†, M. MENDES*, V. PASCHOS* and K. H. NICOLAIDES* *Harris Birthright Research Centre for Fetal Medicine, King’s College Hospital, London, UK; †Department of Fetal Medicine, Medway Maritime Hospital, Gillingham, UK KEYWORDS: first-trimester screening; placental growth factor; pyramid of pregnancy care; stillbirth ABSTRACT the performance of screening achieved by other maternal factors and biomarkers. Copyright © 2016 ISUOG. Objectives To investigate whether the addition of Published by John Wiley & Sons Ltd. maternal serum placental growth factor (PlGF) measured at 11–13 weeks’ gestation improves the performance of screening for stillbirths that is achieved by a combination INTRODUCTION of maternal factors and first-trimester biomarkers such as maternal serum pregnancy-associated plasma protein-A Antepartum stillbirths can be classified broadly into those (PAPP-A), fetal ductus venosus pulsatility index for veins thought to be the consequence of impaired placentation, (DV-PIV) and uterine artery pulsatility index (UtA-PI) including those associated with pre-eclampsia (PE), birth and to evaluate the performance of screening with this of a small-for-gestational-age (SGA) neonate or placental model for all stillbirths and those due to impaired abruption, and those due to other or unexplained causes; placentation and unexplained causes. the rationale of categorizing stillbirths according to the likely underlying cause is that antenatal interventions Methods This was a prospective screening study of and preventive strategies could potentially be undertaken 45 452 singleton pregnancies including 45 225 live births more effectively1–3.