DNA Structure: A-, B- and Z-DNA Helix Families Left-Handed Orientation of the Helix, As Shown in Figure 1B
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

How Can We Create Environments Where Hate Cannot Flourish?
How can we create environments where hate cannot flourish? Saturday, February 1 9:30 a.m. – 10:00 a.m. Check-In and Registration Location: Field Museum West Entrance 10:00 a.m. – 10:30 a.m. Introductions and Program Kick-Off JoAnna Wasserman, USHMM Education Initiatives Manager Location: Lecture Hall 1 10:30 a.m. – 11:30 a.m. Watch “The Path to Nazi Genocide” and Reflections JoAnna Wasserman, USHMM Education Initiatives Manager Location: Lecture Hall 1 11:30 a.m. – 11:45 a.m. Break and walk to State of Deception 11:45 a.m. – 12:30 p.m. Visit State of Deception Interpretation by Holocaust Survivor Volunteers from the Illinois Holocaust Museum Location: Upper level 12:30 p.m. – 1:00 p.m. Breakout Session: Reflections on Exhibit Tim Kaiser, USHMM Director, Education Initiatives David Klevan, USHMM Digital Learning Strategist JoAnna Wasserman, USHMM Education Initiatives Manager Location: Lecture Hall 1, Classrooms A and B Saturday, February 2 (continued) 1:00 p.m. – 1:45 p.m. Lunch 1:45 p.m. – 2:45 p.m. A Survivor’s Personal Story Bob Behr, USHMM Survivor Volunteer Interviewed by: Ann Weber, USHMM Program Coordinator Location: Lecture Hall 1 2:45 p.m. – 3:00 p.m. Break 3:00 p.m. – 3:45 p.m. Student Panel: Beyond Indifference Location: Lecture Hall 1 Moderator: Emma Pettit, Sustained Dialogue Campus Network Student/Alumni Panelists: Jazzy Johnson, Northwestern University Mary Giardina, The Ohio State University Nory Kaplan-Kelly, University of Chicago 3:45 p.m. – 4:30 p.m. Breakout Session: Sharing Personal Reflections Tim Kaiser, USHMM Director, Education Initiatives David Klevan, USHMM Digital Learning Strategist JoAnna Wasserman, USHMM Education Initiatives Manager Location: Lecture Hall 1, Classrooms A and B 4:30 p.m. -

Form 1095-B Health Coverage Department of the Treasury ▶ Do Not Attach to Your Tax Return
560118 VOID OMB No. 1545-2252 Form 1095-B Health Coverage Department of the Treasury ▶ Do not attach to your tax return. Keep for your records. CORRECTED 2020 Internal Revenue Service ▶ Go to www.irs.gov/Form1095B for instructions and the latest information. Part I Responsible Individual 1 Name of responsible individual–First name, middle name, last name 2 Social security number (SSN) or other TIN 3 Date of birth (if SSN or other TIN is not available) 4 Street address (including apartment no.) 5 City or town 6 State or province 7 Country and ZIP or foreign postal code 9 Reserved 8 Enter letter identifying Origin of the Health Coverage (see instructions for codes): . ▶ Part II Information About Certain Employer-Sponsored Coverage (see instructions) 10 Employer name 11 Employer identification number (EIN) 12 Street address (including room or suite no.) 13 City or town 14 State or province 15 Country and ZIP or foreign postal code Part III Issuer or Other Coverage Provider (see instructions) 16 Name 17 Employer identification number (EIN) 18 Contact telephone number 19 Street address (including room or suite no.) 20 City or town 21 State or province 22 Country and ZIP or foreign postal code Part IV Covered Individuals (Enter the information for each covered individual.) (a) Name of covered individual(s) (b) SSN or other TIN (c) DOB (if SSN or other (d) Covered (e) Months of coverage First name, middle initial, last name TIN is not available) all 12 months Jan Feb Mar Apr May Jun Jul Aug Sep Oct Nov Dec 23 24 25 26 27 28 For Privacy Act and Paperwork Reduction Act Notice, see separate instructions. -

State of New Physics in B->S Transitions
New physics in b ! s transitions after LHC run 1 Wolfgang Altmannshofera and David M. Straubb a Perimeter Institute for Theoretical Physics, 31 Caroline St. N, Waterloo, Ontario, Canada N2L 2Y5 b Excellence Cluster Universe, TUM, Boltzmannstr. 2, 85748 Garching, Germany E-mail: [email protected], [email protected] We present results of global fits of all relevant experimental data on rare b s decays. We observe significant tensions between the Standard Model predictions! and the data. After critically reviewing the possible sources of theoretical uncer- tainties, we find that within the Standard Model, the tensions could be explained if there are unaccounted hadronic effects much larger than our estimates. Assuming hadronic uncertainties are estimated in a sufficiently conservative way, we discuss the implications of the experimental results on new physics, both model indepen- dently as well as in the context of the minimal supersymmetric standard model and models with flavour-changing Z0 bosons. We discuss in detail the violation of lepton flavour universality as hinted by the current data and make predictions for additional lepton flavour universality tests that can be performed in the future. We find that the ratio of the forward-backward asymmetries in B K∗µ+µ− and B K∗e+e− at low dilepton invariant mass is a particularly sensitive! probe of lepton! flavour universality and allows to distinguish between different new physics scenarios that give the best description of the current data. Contents 1. Introduction2 2. Observables and uncertainties3 2.1. Effective Hamiltonian . .4 2.2. B Kµ+µ− .....................................4 2.3. B ! K∗µ+µ− and B K∗γ ............................6 ! + − ! 2.4. -

An Introduction to Recurrent Nucleotide Interactions in RNA Blake A
Overview An introduction to recurrent nucleotide interactions in RNA Blake A. Sweeney,1 Poorna Roy2 and Neocles B. Leontis2∗ RNA secondary structure diagrams familiar to molecular biologists summarize at a glance the folding of RNA chains to form Watson–Crick paired double helices. However, they can be misleading: First of all, they imply that the nucleotides in loops and linker segments, which can amount to 35% to 50% of a structured RNA, do not significantly interact with other nucleotides. Secondly, they give the impression that RNA molecules are loosely organized in three-dimensional (3D) space. In fact, structured RNAs are compactly folded as a result of numerous long-range, sequence-specific interactions, many of which involve loop or linker nucleotides. Here, we provide an introduction for students and researchers of RNA on the types, prevalence, and sequence variations of inter-nucleotide interactions that structure and stabilize RNA 3D motifs and architectures, using Escherichia coli (E. coli) 16S ribosomal RNA as a concrete example. The picture that emerges is that almost all nucleotides in structured RNA molecules, including those in nominally single-stranded loop or linker regions, form specific interactions that stabilize functional structures or mediate interactions with other molecules. The small number of noninteracting, ‘looped-out’ nucleotides make it possible for the RNA chain to form sharp turns. Base-pairing is the most specific interaction in RNA as it involves edge-to-edge hydrogen bonding (H-bonding) of the bases. Non-Watson–Crick base pairs are a significant fraction (30% or more) of base pairs in structured RNAs. © 2014 John Wiley & Sons, Ltd. -

Grading System the Grades of A, B, C, D and P Are Passing Grades
Grading System The grades of A, B, C, D and P are passing grades. Grades of F and U are failing grades. R and I are interim grades. Grades of W and X are final grades carrying no credit. Individual instructors determine criteria for letter grade assignments described in individual course syllabi. Explanation of Grades The quality of performance in any academic course is reported by a letter grade, assigned by the instructor. These grades denote the character of study and are assigned quality points as follows: A Excellent 4 grade points per credit B Good 3 grade points per credit C Average 2 grade points per credit D Poor 1 grade point per credit F Failure 0 grade points per credit I Incomplete No credit, used for verifiable, unavoidable reasons. Requirements for satisfactory completion are established through student/faculty consultation. Courses for which the grade of I (incomplete) is awarded must be completed by the end of the subsequent semester or another grade (A, B, C, D, F, W, P, R, S and U) is awarded by the instructor based upon completed course work. In the case of I grades earned at the end of the spring semester, students have through the end of the following fall semester to complete the requirements. In exceptional cases, extensions of time needed to complete course work for I grades may be granted beyond the subsequent semester, with the written approval of the vice president of learning. An I grade can change to a W grade only under documented mitigating circumstances. The vice president of learning must approve the grade change. -
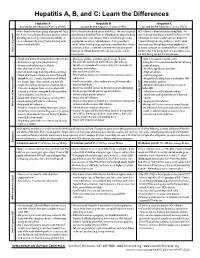
Hepatitis A, B, and C: Learn the Differences
Hepatitis A, B, and C: Learn the Differences Hepatitis A Hepatitis B Hepatitis C caused by the hepatitis A virus (HAV) caused by the hepatitis B virus (HBV) caused by the hepatitis C virus (HCV) HAV is found in the feces (poop) of people with hepa- HBV is found in blood and certain body fluids. The virus is spread HCV is found in blood and certain body fluids. The titis A and is usually spread by close personal contact when blood or body fluid from an infected person enters the body virus is spread when blood or body fluid from an HCV- (including sex or living in the same household). It of a person who is not immune. HBV is spread through having infected person enters another person’s body. HCV can also be spread by eating food or drinking water unprotected sex with an infected person, sharing needles or is spread through sharing needles or “works” when contaminated with HAV. “works” when shooting drugs, exposure to needlesticks or sharps shooting drugs, through exposure to needlesticks on the job, or from an infected mother to her baby during birth. or sharps on the job, or sometimes from an infected How is it spread? Exposure to infected blood in ANY situation can be a risk for mother to her baby during birth. It is possible to trans- transmission. mit HCV during sex, but it is not common. • People who wish to be protected from HAV infection • All infants, children, and teens ages 0 through 18 years There is no vaccine to prevent HCV. -

I/B/E/S DETAIL HISTORY a Guide to the Analyst-By-Analyst Historical Earnings Estimate Database
I/B/E/S DETAIL HISTORY A guide to the analyst-by-analyst historical earnings estimate database U.S. Edition CONTENTS page Overview..................................................................................................................................................................................................................1 File Explanations ..............................................................................................................................................................................................2 Detail File ..................................................................................................................................................................................................2 Identifier File..........................................................................................................................................................................................3 Adjustments File..................................................................................................................................................................................3 Excluded Estimates File ...............................................................................................................................................................3 Broker Translations...........................................................................................................................................................................4 S/I/G Codes..............................................................................................................................................................................................4 -

Alphabets, Letters and Diacritics in European Languages (As They Appear in Geography)
1 Vigleik Leira (Norway): [email protected] Alphabets, Letters and Diacritics in European Languages (as they appear in Geography) To the best of my knowledge English seems to be the only language which makes use of a "clean" Latin alphabet, i.d. there is no use of diacritics or special letters of any kind. All the other languages based on Latin letters employ, to a larger or lesser degree, some diacritics and/or some special letters. The survey below is purely literal. It has nothing to say on the pronunciation of the different letters. Information on the phonetic/phonemic values of the graphic entities must be sought elsewhere, in language specific descriptions. The 26 letters a, b, c, d, e, f, g, h, i, j, k, l, m, n, o, p, q, r, s, t, u, v, w, x, y, z may be considered the standard European alphabet. In this article the word diacritic is used with this meaning: any sign placed above, through or below a standard letter (among the 26 given above); disregarding the cases where the resulting letter (e.g. å in Norwegian) is considered an ordinary letter in the alphabet of the language where it is used. Albanian The alphabet (36 letters): a, b, c, ç, d, dh, e, ë, f, g, gj, h, i, j, k, l, ll, m, n, nj, o, p, q, r, rr, s, sh, t, th, u, v, x, xh, y, z, zh. Missing standard letter: w. Letters with diacritics: ç, ë. Sequences treated as one letter: dh, gj, ll, rr, sh, th, xh, zh. -
![Building the B[R]And: Understanding How Social Media Drives Consumer Engagement and Sales](https://docslib.b-cdn.net/cover/1301/building-the-b-r-and-understanding-how-social-media-drives-consumer-engagement-and-sales-801301.webp)
Building the B[R]And: Understanding How Social Media Drives Consumer Engagement and Sales
Marketing Science Institute Working Paper Series 2013 Report No. 13-113 Building the B[r]and: Understanding How Social Media Drives Consumer Engagement and Sales Yogesh V. Joshi , Liye Ma, William M. Rand, and Louiqa Raschid “Building the B[r]and: Understanding How Social Media Drives Consumer Engagement and Sales” © 2013 Yogesh V. Joshi , Liye Ma, William M. Rand, and Louiqa Raschid; Report Summary © 2013 Marketing Science Institute MSI working papers are distributed for the benefit of MSI corporate and academic members and the general public. Reports are not to be reproduced or published in any form or by any means, electronic or mechanical, without written permission. Report Summary Yogesh Joshi , Liye Ma, William Rand, and Louiqa Raschid investigate how activity in digital social media, by new and established brands, relates to engagement with consumers, and eventually, sales. Their dataset includes two years of Twitter activity and offline concerts for several musical bands, and the corresponding social media activity of the bands’ followers. In addition to measuring volume (that is, number of tweets sent per unit of time), the authors use machine learning methods to analyze message sentiment and informational content. They also collect A.C. Nielsen sales data for all albums released by these bands. They investigate the characteristics and evolution of consumers' engagement (propensity to tweet in response to a band’s tweets as well as propensity to send informational or emotional tweets) using a hidden Markov model. They relate engagement to sales via a generalized diffusion model. Findings Overall, the authors find that band actions in social media generate interest and change in engagement levels of their followers, and these engagement levels have a positive association with sales. -

Review Article Use of Nucleic Acid Analogs for the Study of Nucleic Acid Interactions
SAGE-Hindawi Access to Research Journal of Nucleic Acids Volume 2011, Article ID 967098, 11 pages doi:10.4061/2011/967098 Review Article Use of Nucleic Acid Analogs for the Study of Nucleic Acid Interactions Shu-ichi Nakano,1, 2 Masayuki Fujii,3, 4 and Naoki Sugimoto1, 2 1 Faculty of Frontiers of Innovative Research in Science and Technology, Konan University, 7-1-20 Minatojima-Minamimachi, Chuo-ku, Kobe 650-0047, Japan 2 Frontier Institute for Biomolecular Engineering Research, Konan University, 7-1-20 Minatojima-Minamimachi, Chuo-ku, Kobe 650-0047, Japan 3 Department of Environmental and Biological Chemistry, Kinki University, 11-6 Kayanomori, Iizuka, Fukuoka 820-8555, Japan 4 Molecular Engineering Institute, Kinki University, 11-6 Kayanomori, Iizuka, Fukuoka 820-8555, Japan Correspondence should be addressed to Shu-ichi Nakano, [email protected] and Naoki Sugimoto, [email protected] Received 14 April 2011; Accepted 2 May 2011 Academic Editor: Daisuke Miyoshi Copyright © 2011 Shu-ichi Nakano et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Unnatural nucleosides have been explored to expand the properties and the applications of oligonucleotides. This paper briefly summarizes nucleic acid analogs in which the base is modified or replaced by an unnatural stacking group for the study of nucleic acid interactions. We also describe the nucleoside analogs of a base pair-mimic structure that we have examined. Although the base pair-mimic nucleosides possess a simplified stacking moiety of a phenyl or naphthyl group, they can be used as a structural analog of Watson-Crick base pairs. -

Questions with Answers- Nucleotides & Nucleic Acids A. the Components
Questions with Answers- Nucleotides & Nucleic Acids A. The components and structures of common nucleotides are compared. (Questions 1-5) 1._____ Which structural feature is shared by both uracil and thymine? a) Both contain two keto groups. b) Both contain one methyl group. c) Both contain a five-membered ring. d) Both contain three nitrogen atoms. 2._____ Which component is found in both adenosine and deoxycytidine? a) Both contain a pyranose. b) Both contain a 1,1’-N-glycosidic bond. c) Both contain a pyrimidine. d) Both contain a 3’-OH group. 3._____ Which property is shared by both GDP and AMP? a) Both contain the same charge at neutral pH. b) Both contain the same number of phosphate groups. c) Both contain the same purine. d) Both contain the same furanose. 4._____ Which characteristic is shared by purines and pyrimidines? a) Both contain two heterocyclic rings with aromatic character. b) Both can form multiple non-covalent hydrogen bonds. c) Both exist in planar configurations with a hemiacetal linkage. d) Both exist as neutral zwitterions under cellular conditions. 5._____ Which property is found in nucleosides and nucleotides? a) Both contain a nitrogenous base, a pentose, and at least one phosphate group. b) Both contain a covalent phosphodister bond that is broken in strong acid. c) Both contain an anomeric carbon atom that is part of a β-N-glycosidic bond. d) Both contain an aldose with hydroxyl groups that can tautomerize. ___________________________________________________________________________ B. The structures of nucleotides and their components are studied. (Questions 6-10) 6._____ Which characteristic is shared by both adenine and cytosine? a) Both contain one methyl group. -

Proposal for Generation Panel for Latin Script Label Generation Ruleset for the Root Zone
Generation Panel for Latin Script Label Generation Ruleset for the Root Zone Proposal for Generation Panel for Latin Script Label Generation Ruleset for the Root Zone Table of Contents 1. General Information 2 1.1 Use of Latin Script characters in domain names 3 1.2 Target Script for the Proposed Generation Panel 4 1.2.1 Diacritics 5 1.3 Countries with significant user communities using Latin script 6 2. Proposed Initial Composition of the Panel and Relationship with Past Work or Working Groups 7 3. Work Plan 13 3.1 Suggested Timeline with Significant Milestones 13 3.2 Sources for funding travel and logistics 16 3.3 Need for ICANN provided advisors 17 4. References 17 1 Generation Panel for Latin Script Label Generation Ruleset for the Root Zone 1. General Information The Latin script1 or Roman script is a major writing system of the world today, and the most widely used in terms of number of languages and number of speakers, with circa 70% of the world’s readers and writers making use of this script2 (Wikipedia). Historically, it is derived from the Greek alphabet, as is the Cyrillic script. The Greek alphabet is in turn derived from the Phoenician alphabet which dates to the mid-11th century BC and is itself based on older scripts. This explains why Latin, Cyrillic and Greek share some letters, which may become relevant to the ruleset in the form of cross-script variants. The Latin alphabet itself originated in Italy in the 7th Century BC. The original alphabet contained 21 upper case only letters: A, B, C, D, E, F, Z, H, I, K, L, M, N, O, P, Q, R, S, T, V and X.