Expression of Pgc1α in Glioblastoma Multiforme Patients
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Supplementary Table 1
Supplementary Table 1. 492 genes are unique to 0 h post-heat timepoint. The name, p-value, fold change, location and family of each gene are indicated. Genes were filtered for an absolute value log2 ration 1.5 and a significance value of p ≤ 0.05. Symbol p-value Log Gene Name Location Family Ratio ABCA13 1.87E-02 3.292 ATP-binding cassette, sub-family unknown transporter A (ABC1), member 13 ABCB1 1.93E-02 −1.819 ATP-binding cassette, sub-family Plasma transporter B (MDR/TAP), member 1 Membrane ABCC3 2.83E-02 2.016 ATP-binding cassette, sub-family Plasma transporter C (CFTR/MRP), member 3 Membrane ABHD6 7.79E-03 −2.717 abhydrolase domain containing 6 Cytoplasm enzyme ACAT1 4.10E-02 3.009 acetyl-CoA acetyltransferase 1 Cytoplasm enzyme ACBD4 2.66E-03 1.722 acyl-CoA binding domain unknown other containing 4 ACSL5 1.86E-02 −2.876 acyl-CoA synthetase long-chain Cytoplasm enzyme family member 5 ADAM23 3.33E-02 −3.008 ADAM metallopeptidase domain Plasma peptidase 23 Membrane ADAM29 5.58E-03 3.463 ADAM metallopeptidase domain Plasma peptidase 29 Membrane ADAMTS17 2.67E-04 3.051 ADAM metallopeptidase with Extracellular other thrombospondin type 1 motif, 17 Space ADCYAP1R1 1.20E-02 1.848 adenylate cyclase activating Plasma G-protein polypeptide 1 (pituitary) receptor Membrane coupled type I receptor ADH6 (includes 4.02E-02 −1.845 alcohol dehydrogenase 6 (class Cytoplasm enzyme EG:130) V) AHSA2 1.54E-04 −1.6 AHA1, activator of heat shock unknown other 90kDa protein ATPase homolog 2 (yeast) AK5 3.32E-02 1.658 adenylate kinase 5 Cytoplasm kinase AK7 -

Systems Physiology and Nutrition In
SYSTEMS PHYSIOLOGY AND NUTRITION IN DAIRY CATTLE: APPLICATIONS OF OMICS AND BIOINFORMATICS TO BETTER UNDERSTAND THE HEPATIC METABOLOMICS AND TRANSCRIPTOMICS ADAPTATIONS IN TRANSITION DAIRY COWS BY KHURAM SHAHZAD DISSERTATION Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Informatics in the Graduate College of the University of Illinois at Urbana-Champaign, 2017 Urbana, Illinois Doctoral Committee: Associate Professor, Juan J. Loor, Chair Professor Gustavo Caetano-Anolles Associate Professor, Juan Steibel, Michigan State University Assistant Professor Phil Cardoso ABSTRACT Application of systems concepts to better understand physiological and metabolic changes in dairy cows during the transition into lactation could enhance our understanding about the role of nutrients in helping to meet the animal’s requirements for optimal production and health. Four different analyses focused on the liver were conducted to analyze metabolic disorder or thermal stress. The first three analyses dealt with supplementation of methionine to prevent clinical ketosis development in high-genetic merit dairy cows. Four groups of cows were formed retrospectively based on clinical health evaluated at 1 week postpartum: cows that remained healthy (OVE), cows that developed ketosis (K), and healthy cows supplemented with one of two commercial methionine products [Smartamine M (SM), and MetaSmart (MS)]. The liver tissue samples (n = 6/group) were harvested at -10 d before calving, and were used for metabolomics (GC-MS, LC-MS; Metabolon Inc.) and transcriptomics (44K-whole-transcriptome microarray; Agilent) analyses. Therefore, the main goals of the analyses were to 1) uncover metabolome and transcriptome patterns in the prepartum liver that were unique to those cows that became ketotic postpartum, and to 2) uncover unique patterns affected by supplemental methionine. -

WO 2013/184908 A2 12 December 2013 (12.12.2013) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization I International Bureau (10) International Publication Number (43) International Publication Date WO 2013/184908 A2 12 December 2013 (12.12.2013) P O P C T (51) International Patent Classification: Jr.; One Procter & Gamble Plaza, Cincinnati, Ohio 45202 G06F 19/00 (201 1.01) (US). HOWARD, Brian, Wilson; One Procter & Gamble Plaza, Cincinnati, Ohio 45202 (US). (21) International Application Number: PCT/US20 13/044497 (74) Agents: GUFFEY, Timothy, B. et al; c/o The Procter & Gamble Company, Global Patent Services, 299 East 6th (22) Date: International Filing Street, Sycamore Building, 4th Floor, Cincinnati, Ohio 6 June 2013 (06.06.2013) 45202 (US). (25) Filing Language: English (81) Designated States (unless otherwise indicated, for every (26) Publication Language: English kind of national protection available): AE, AG, AL, AM, AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, (30) Priority Data: BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, 61/656,218 6 June 2012 (06.06.2012) US DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, (71) Applicant: THE PROCTER & GAMBLE COMPANY HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KN, KP, KR, [US/US]; One Procter & Gamble Plaza, Cincinnati, Ohio KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, ME, 45202 (US). MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SC, (72) Inventors: XU, Jun; One Procter & Gamble Plaza, Cincin SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, nati, Ohio 45202 (US). -

Distribution Agreement
Distribution Agreement In presenting this thesis or dissertation as a partial fulfillment of the requirements for an advanced degree from Emory University, I hereby grant to Emory University and its agents the non-exclusive license to archive, make accessible, and display my thesis or dissertation in whole or in part in all forms of media, now or hereafter known, including display on the world wide web. I understand that I may select some access restrictions as part of the online submission of this thesis or dissertation. I retain all ownership rights to the copyright of the thesis or dissertation. I also retain the right to use in future works (such as articles or books) all or part of this thesis or dissertation. Signature: _____________________________ ______________ Ivan Ludlow Date MDMA’s Effect on Myocardial DNA Methylation and its Association with Dilated Cardiomyopathy By Ivan Ludlow Master of Public Health Environmental Health _________________________________________ William Michael Caudle, Ph.D. Committee Chair _________________________________________ Paige Tolbert, Ph.D. Committee Member _________________________________________ William Lewis, M.D. Committee Member MDMA’s Effect on Myocardial DNA Methylation and its Association with Dilated Cardiomyopathy By Ivan Ludlow B.S. Emory University 2014 Thesis Committee Chair: William Michael Caudle, Ph.D. An abstract of A thesis submitted to the Faculty of the Rollins School of Public Health of Emory University in partial fulfillment of the requirements for the degree of Master of Public Health in Environmental Health 2015 Abstract MDMA’s Effect on Myocardial DNA Methylation and its Association with Dilated Cardiomyopathy By Ivan Ludlow MDMA (“Ecstasy”) is an illicit psychoactive drug that has increased in popularity in the past two decades. -
B a C D E F G K
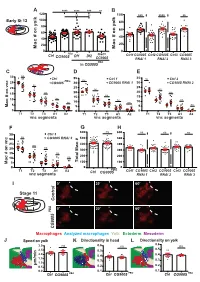
A **** **** *** ns B 120 ns ns 100 *** **** ** 100 Early St 12 80 80 60 on yolk 60 on yolk # # 40 40 ac 20 ac 20 M 0 M 0 PBG mac> Ctrl CG9005 Df1 Df2 Ctrl1CG9005 Ctrl2 CG9005 Ctrl3 CG9005 CG9005 RNAi 1 RNAi 2 RNAi 3 PBG in CG9005 CDE 30 ns Ctrl 35 Ctrl 1 35 ns Ctrl 2 PBG ns 25 CG9005 30 CG9005 RNAi 1 30 CG9005 RNAi 2 ns 25 25 20 ns ns ns 20 20 ns on vnc 15 ns 15 # ns 15 10 ns ns ns 10 10 ac ns ns 5 5 5 M 0 0 0 T1 T2 T3 A1 A2 T1 T2 T3 A1 A2 T1 T2 T3 A1 A2 vnc segments vnc segments vnc segments F G H 600 ns 600 ns ns ns 35 Ctrl 3 ns 30 CG9005 RNAi 3 500 500 # 25 ns 400 400 ns ac 20 300 300 on vnc 15 ns # 200 200 10 ns ac 100 5 Total M 100 M 0 0 0 PBG T1 T2 T3 A1 A2 Ctrl CG9005 Ctrl1CG9005 Ctrl2 CG9005 Ctrl3 CG9005 vnc segments RNAi 1 RNAi 2 RNAi 3 I 0’ 30’ 60’ Stage 11 Control 0’ 30’ 60’ PBG CG9005 Macrophages Analyzed macrophages Yolk Ectoderm Mesoderm J Speed on yolk K Directionality in head L Directionality on yolk 0.6 3.0 ns 0.6 ns ns 0.5 2.5 0.5 in 2.0 0.4 0.4 1.5 0.3 0.3 m/m 1.0 μ 0.2 0.5 0.2 0.0 0.1 0.1 PBG PBG Ctrl CG9005 PBG Ctrl CG9005 Ctrl CG9005 Figure S1 related to Figure 1: CG9005PBG mutant macrophages migrate normally within the head and along the vnc. -

A Systems Chemical Biology Approach for Dissecting Differential
University of South Florida Scholar Commons Graduate Theses and Dissertations Graduate School June 2018 A Systems Chemical Biology Approach for Dissecting Differential Molecular Mechanisms of Action of Clinical Kinase Inhibitors in Lung Cancer Natalia Junqueira Sumi University of South Florida, [email protected] Follow this and additional works at: https://scholarcommons.usf.edu/etd Part of the Molecular Biology Commons Scholar Commons Citation Junqueira Sumi, Natalia, "A Systems Chemical Biology Approach for Dissecting Differential Molecular Mechanisms of Action of Clinical Kinase Inhibitors in Lung Cancer" (2018). Graduate Theses and Dissertations. https://scholarcommons.usf.edu/etd/7685 This Dissertation is brought to you for free and open access by the Graduate School at Scholar Commons. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Scholar Commons. For more information, please contact [email protected]. A Systems Chemical Biology Approach for Dissecting Differential Molecular Mechanisms of Action of Clinical Kinase Inhibitors in Lung Cancer by Natalia Junqueira Sumi A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy with a concentration in Cancer Biology Department of Cell Biology, Microbiology and Molecular Biology College of Arts and Sciences University of South Florida Major Professor: Uwe Rix, Ph.D. Nicholas J. Lawrence, Ph.D. Keiran S. Smalley, Ph.D. W. Douglas Cress, Ph.D. Eric B. Haura, MD. Forest White, PhD. Date of Approval: March 31, 2018 Keywords: Proteomics, lung cancer, RNA-seq, network, bioinformatics, drug combination Copyright © 2018, Natalia Junqueira Sumi DEDICATION This dissertation is primarily dedicated to me. -

Gene Expression Profiling at Birth Characterizing the Preterm Infant with Early Onset Infection
J Mol Med (2017) 95:169–180 DOI 10.1007/s00109-016-1466-4 ORIGINAL ARTICLE Gene expression profiling at birth characterizing the preterm infant with early onset infection Anne Hilgendorff1,2 & Anita Windhorst 3,4 & Manuel Klein5 & Svetlin Tchatalbachev3 & Christine Windemuth-Kieselbach6 & Joachim Kreuder2 & Matthias Heckmann7 & Anna Gkatzoflia2 & Harald Ehrhardt2 & Josef Mysliwietz8 & Michael Maier3 & Benjamin Izar3,9 & Andre Billion3 & Ludwig Gortner10 & Trinad Chakraborty3 & Hamid Hossain3 Received: 26 August 2015 /Revised: 9 August 2016 /Accepted: 18 August 2016 /Published online: 30 August 2016 # The Author(s) 2016. This article is published with open access at Springerlink.com Abstract with EOI could be further differentiated into two subclasses Early onset infection (EOI) in preterm infants <32 weeks ges- and were distinguished by the magnitude of the expression of tational age (GA) is associated with a high mortality rate and genes involved in both neutrophil and T cell activation. A the development of severe acute and long-term complications. hallmark activity for both subclasses of EOI was a common The pathophysiology of EOI is not fully understood and clin- suppression of genes involved in natural killer (NK) cell func- ical and laboratory signs of early onset infections in this pa- tion, which was independent from NK cell numbers. tient cohort are often not conclusive. Thus, the aim of this Significant results were recapitulated in an independent vali- study was to identify signatures characterizing preterm infants dation cohort. Gene expression profiling may enable early and with EOI by using genome-wide gene expression (GWGE) more precise diagnosis of EOI in preterm infants. analyses from umbilical arterial blood of preterm infants. -

Identification of Liver Metastasis-Associated Genes in Human Colon Carcinoma by Mrna Profiling
Original Article Identification of liver metastasis-associated genes in human colon carcinoma by mRNA profiling Jianling Liu1,2*, Dan Wang1,2*, Chaoqi Zhang1,2*, Zhen Zhang1,2, Xinfeng Chen1,2, Jingyao Lian1,2, Jinbo Liu4, Guixian Wang4, Weitang Yuan4, Zhenqiang Sun4, Weijia Wang1,2, Mengjia Song1,2, Yaping Wang5, Qian Wu1,2, Ling Cao1,2, Dong Wang1,2, Yi Zhang1,2,3 1Biotherapy Center, the First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, China; 2Department of Oncology, the First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, China; 3School of Life Sciences, Zhengzhou University, Zhengzhou 450001, China; 4Department of Anorectal Surgery, the First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, China; 5Henan Key Laboratory for Tumor Immunology and Biotherapy, Zhengzhou 450052, China *These authors contributed equally to this work. Correspondence to: Yi Zhang. Biotherapy Center, the First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, China. Email: [email protected]. Abstract Objective: Liver metastasis, which contributes substantially to high mortality, is the most common recurrent mode of colon carcinoma. Thus, it is necessary to identify genes implicated in metastatic colonization of the liver in colon carcinoma. Methods: We compared mRNA profiling in 18 normal colon mucosa (N), 20 primary tumors (T) and 19 liver metastases (M) samples from the dataset GSE49355 and GSE62321 of Gene Expression Omnibus (GEO) database. Gene ontology (GO) and pathways of the identified genes were analyzed. Co-expression network and protein- protein interaction (PPI) network were employed to identify the interaction relationship. Survival analyses based on The Cancer Genome Atlas (TCGA) database were used to further screening. -

STO (%) % O (STO) Slices of 12Μm Thickness After Equilibration to -20 C
Table S1 Definition and method of traits Traits Definitions1 Unit Methods 2,3 Percentage of slow-twitch-oxidative fiber The cryopreserved muscle samples were cutting into STO (%) % o (STO) slices of 12µm thickness after equilibration to -20 C. NADH tetrazolium reductase and Myofibrillar ATPase Percentage of fast-twitch-oxidative fiber FTO (%) % were stained to identify the muscle fiber types. 3 sections (FTO) were used for calculating the percentage of STO, FTO Percentage of fast-twitch-glycolytic fiber and FTG by relating the number of counted fibers of each FTG (%) % (FTG) type to the total counted fiber number. State 3 mitochondrial respiratory activity pmol State 3 (MRA) analyzed with substrate combination O2/sec*mg 2 Pyruvate Samples were dissected and muscle fibers were pyruvate/malate sample weight mechanically permeabilized, dried on filter paper and State 3 mitochondrial respiratory activity pmol State 3 weighted. The MRA was measured using Oxygraph (MRA) analyzed with substrate combination O2/sec*mg Succinate equipped with a Clark-electrode. The weight-specific succinate/rotenone sample weight oxygen consumption (pmol O2 /sec*mg sample weight) pmol was calculated as the time derivative of oxygen State 4 State 4 mitochondrial respiratory activity O2/sec*mg concentration. State 3 respiration was initiated with 5mM CAT initiated with carboxy-atractyloside (CAT) sample weight ADP. State 4 respiration was initiated with 28 µM CAT. RCI Respiratory control index (RCI) when RCI Pyruvate was calculated by dividing the state 3 no Pyruvate considering substrates pyruvate/malate Pyruvate and state 4 respiration rate. Each experiment RCI Respiratory control index (RCI) when was repeated three times. -

MF) Using Isobaric Tags for Relative and Absolute Quantification (Itraq
Hindawi BioMed Research International Volume 2020, Article ID 9237381, 13 pages https://doi.org/10.1155/2020/9237381 Research Article Proteomic Profiling Change and Its Implies in the Early Mycosis Fungoides (MF) Using Isobaric Tags for Relative and Absolute Quantification (iTRAQ) Mengyan Zhu ,1 Yong Li ,2,3,4 Cheng Ding ,5 Jiaqi Wang ,1 Yangyang Ma ,2 Zhao Li ,2 Xiaoyan Zhang ,2 and Ping Wang 2 1The Third People’s Hospital of Hangzhou Affiliated to Zhejiang Chinese Medical University, Hangzhou 310009, China 2The Third People’s Hospital of Hangzhou, Hangzhou 310009, China 3Institute of Plant Physiology and Ecology, SIBS, CAS, Shanghai, China 4Research Center, Shanghai Yeslab Biotechnology, Shanghai, China 5People’s Hospital of Cangzhou, Hebei, China Correspondence should be addressed to Ping Wang; [email protected] Received 16 July 2020; Revised 1 October 2020; Accepted 31 October 2020; Published 24 November 2020 Academic Editor: Nobuo Kanazawa Copyright © 2020 Mengyan Zhu et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Purpose. Mycosis fungoides (MF) is the most common T-cell lymphoma, with indolent biologic behavior in the early stage and features of invasive in the tumor stage. The diagnosis of MF is still ambiguous and difficult. We focused on the proteomic profiling change in the pathogenesis of early MF and identified candidate biomarkers for early diagnosis. Methods. We collected peripheral blood samples of MF patients and healthy individuals (HI) performed proteomic profiling analysis using isobaric tags for relative and absolute quantification (iTRAQ) platform. -

1 KLF2 and KLF4 Control Endothelial Identity and Vascular Integrity
KLF2 and KLF4 control endothelial identity and vascular integrity Panjamaporn Sangwung 1, 2,#, Guangjin Zhou1,#, Lalitha Nayak1,3, E. Ricky Chan4, Sandeep Kumar5, Dong-Won Kang5, Rongli Zhang1, Xudong Liao1, Yuan Lu1, Keiki Sugi1, Hisashi Fujioka7, Hong Shi1, Stephanie D Lapping1, Chandra C. Ghosh8, Sarah J. Higgins8, Samir M. Parikh8, Hanjoon Jo5,6, and Mukesh K Jain1,2,9 Supplemental Figures and Tables 1 Supplemental Figure 1 A Klf2 Klf4 Nos3 Thbd 3 ** 3 ** 3 ** 3 ** NS ** * NS ** NS * NS Day 6 2 2 2 2 post-tamoxifen 1 1 1 1 expression Relative mRNA Relative mRNA 0 0 0 0 CRE -KO -KO CRE -KO -KO CRE -KO -KO CRE -KO -KO Klf2 Klf4 EC-DKO Klf2 Klf4 EC-DKO Klf2 Klf4 EC-DKO Klf2 Klf4 EC-DKO EC- EC- EC- EC- EC- EC- EC- EC- B Klf2 Klf4 Nos3 Thbd 3 ** 3 ** 3 ** 3 ** NS Day 5 ** ** ** ** NS NS * post-tamoxifen 2 2 2 2 1 1 1 1 expression Relative mRNA Relative mRNA 0 0 0 0 CRE -KO -KO CRE -KO -KO CRE -KO -KO CRE -KO -KO Klf2 Klf4 EC-DKO Klf2 Klf4 EC-DKO Klf2 Klf4 EC-DKO Klf2 Klf4 EC-DKO EC- EC- EC- EC- EC- EC- EC- EC- C Klf2 Klf4 Nos3 Thbd 3 ** 3 ** 3 ** 3 ** NS ** NS NS Day 4 ** NS NS NS 2 2 2 2 post-tamoxifen 1 1 1 1 expression Relative mRNA Relative mRNA 0 0 0 0 CRE -KO -KO CRE -KO -KO CRE -KO -KO CRE -KO -KO Klf2 Klf4 EC-DKO Klf2 Klf4 EC-DKO Klf2 Klf4 EC-DKO Klf2 Klf4 EC-DKO EC- EC- EC- EC- EC- EC- EC- EC- D CRE EC-DKO KLF4 TM GAPDH eNOS GAPDH Supplemental Figure 1.