Appendix 1: Reference Database Construction
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Disaggregation of Bird Families Listed on Cms Appendix Ii
Convention on the Conservation of Migratory Species of Wild Animals 2nd Meeting of the Sessional Committee of the CMS Scientific Council (ScC-SC2) Bonn, Germany, 10 – 14 July 2017 UNEP/CMS/ScC-SC2/Inf.3 DISAGGREGATION OF BIRD FAMILIES LISTED ON CMS APPENDIX II (Prepared by the Appointed Councillors for Birds) Summary: The first meeting of the Sessional Committee of the Scientific Council identified the adoption of a new standard reference for avian taxonomy as an opportunity to disaggregate the higher-level taxa listed on Appendix II and to identify those that are considered to be migratory species and that have an unfavourable conservation status. The current paper presents an initial analysis of the higher-level disaggregation using the Handbook of the Birds of the World/BirdLife International Illustrated Checklist of the Birds of the World Volumes 1 and 2 taxonomy, and identifies the challenges in completing the analysis to identify all of the migratory species and the corresponding Range States. The document has been prepared by the COP Appointed Scientific Councilors for Birds. This is a supplementary paper to COP document UNEP/CMS/COP12/Doc.25.3 on Taxonomy and Nomenclature UNEP/CMS/ScC-Sc2/Inf.3 DISAGGREGATION OF BIRD FAMILIES LISTED ON CMS APPENDIX II 1. Through Resolution 11.19, the Conference of Parties adopted as the standard reference for bird taxonomy and nomenclature for Non-Passerine species the Handbook of the Birds of the World/BirdLife International Illustrated Checklist of the Birds of the World, Volume 1: Non-Passerines, by Josep del Hoyo and Nigel J. Collar (2014); 2. -

Thailand Highlights 14Th to 26Th November 2019 (13 Days)
Thailand Highlights 14th to 26th November 2019 (13 days) Trip Report Siamese Fireback by Forrest Rowland Trip report compiled by Tour Leader: Forrest Rowland Trip Report – RBL Thailand - Highlights 2019 2 Tour Summary Thailand has been known as a top tourist destination for quite some time. Foreigners and Ex-pats flock there for the beautiful scenery, great infrastructure, and delicious cuisine among other cultural aspects. For birders, it has recently caught up to big names like Borneo and Malaysia, in terms of respect for the avian delights it holds for visitors. Our twelve-day Highlights Tour to Thailand set out to sample a bit of the best of every major habitat type in the country, with a slight focus on the lush montane forests that hold most of the country’s specialty bird species. The tour began in Bangkok, a bustling metropolis of winding narrow roads, flyovers, towering apartment buildings, and seemingly endless people. Despite the density and throng of humanity, many of the participants on the tour were able to enjoy a Crested Goshawk flight by Forrest Rowland lovely day’s visit to the Grand Palace and historic center of Bangkok, including a fun boat ride passing by several temples. A few early arrivals also had time to bird some of the urban park settings, even picking up a species or two we did not see on the Main Tour. For most, the tour began in earnest on November 15th, with our day tour of the salt pans, mudflats, wetlands, and mangroves of the famed Pak Thale Shore bird Project, and Laem Phak Bia mangroves. -

WAR and PROTECTED AREAS AREAS and PROTECTED WAR Vol 14 No 1 Vol 14 Protected Areas Programme Areas Protected
Protected Areas Programme Protected Areas Programme Vol 14 No 1 WAR AND PROTECTED AREAS 2004 Vol 14 No 1 WAR AND PROTECTED AREAS 2004 Parks Protected Areas Programme © 2004 IUCN, Gland, Switzerland Vol 14 No 1 WAR AND PROTECTED AREAS 2004 ISSN: 0960-233X Vol 14 No 1 WAR AND PROTECTED AREAS CONTENTS Editorial JEFFREY A. MCNEELY 1 Parks in the crossfire: strategies for effective conservation in areas of armed conflict JUDY OGLETHORPE, JAMES SHAMBAUGH AND REBECCA KORMOS 2 Supporting protected areas in a time of political turmoil: the case of World Heritage 2004 Sites in the Democratic Republic of Congo GUY DEBONNET AND KES HILLMAN-SMITH 9 Status of the Comoé National Park, Côte d’Ivoire and the effects of war FRAUKE FISCHER 17 Recovering from conflict: the case of Dinder and other national parks in Sudan WOUTER VAN HOVEN AND MUTASIM BASHIR NIMIR 26 Threats to Nepal’s protected areas PRALAD YONZON 35 Tayrona National Park, Colombia: international support for conflict resolution through tourism JENS BRÜGGEMANN AND EDGAR EMILIO RODRÍGUEZ 40 Establishing a transboundary peace park in the demilitarized zone on the Kuwaiti/Iraqi borders FOZIA ALSDIRAWI AND MUNA FARAJ 48 Résumés/Resumenes 56 Subscription/advertising details inside back cover Protected Areas Programme Vol 14 No 1 WAR AND PROTECTED AREAS 2004 ■ Each issue of Parks addresses a particular theme, in 2004 these are: Vol 14 No 1: War and protected areas Vol 14 No 2: Durban World Parks Congress Vol 14 No 3: Global change and protected areas ■ Parks is the leading global forum for information on issues relating to protected area establishment and management ■ Parks puts protected areas at the forefront of contemporary environmental issues, such as biodiversity conservation and ecologically The international journal for protected area managers sustainable development ISSN: 0960-233X Published three times a year by the World Commission on Protected Areas (WCPA) of IUCN – Subscribing to Parks The World Conservation Union. -

Siberian Blue Robin Larvivora Cyane from the Barak Valley of Assam with a Status Update for India
See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/338137338 Siberian Blue Robin Larvivora cyane from the Barak Valley of Assam with a status update for India Article in Indian BIRDS · December 2019 CITATIONS READS 0 7 2 authors: Rejoice Gassah Vijay Anand Ismavel Makunda Christian Hospital 4 PUBLICATIONS 0 CITATIONS 22 PUBLICATIONS 12 CITATIONS SEE PROFILE SEE PROFILE Some of the authors of this publication are also working on these related projects: Neglected health problems in rural India View project Biodiversity Documentation View project All content following this page was uploaded by Vijay Anand Ismavel on 24 December 2019. The user has requested enhancement of the downloaded file. Correspondence 123 peninsular India (Grimmett et al. 2011; Rasmussen & Anderton minY=11.103753465762485&env.maxX=93.01342361450202&env.maxY=12.31963 2012; eBird 2019). This species is a common winter visitor to 3103994705&zh=true&gp=true&ev=Z&mr=on&bmo=1&emo=1&yr=cur&byr=2019 Myanmar, Thailand, Laos, and Tonkin (Robson 2008). &eyr=2019. [Accessed on: 19 January 2019.] DeCandido, R., Subedi, T., Siponen, M., Sutasha, K., Pierce, A., Nualsri, C., & Round, P. D., 2013. Flight identification of Milvus migrans lineatus ‘Black-eared’ Kite and Milvus migrans govinda ‘Pariah’ Kite in Nepal and Thailand. BirdingASIA 20: 32–36. Grimmett, R., Inskipp, C., & Inskipp, T., 2011. Birds of the Indian Subcontinent. 2nd ed. London: Oxford University Press & Christopher Helm. Pp. 1–528. Rasmussen, P. C., & Anderton, J. C., 2012. Birds of South Asia: the Ripley guide: field guide. 2nd ed. Washington, D.C. -

First Images in the Wild of Blackthroat Luscinia Obscura, Asia's Most
BirdingASIA 15 (2011): 17–19 17 LITTLE-KNOWN ASIAN BIRD First images in the wild of Blackthroat Luscinia obscura, Asia’s most enigmatic robin WEI QIAN & HE YI Background probably a female from Baihe Nature Reserve, The Blackthroat Luscinia obscura is known to breed Sichuan, on 3 June 2007 (Anderson 2007). only in the mountains of south-west China, where Additionally, a few captive birds, assumed to have there are a few scattered records from Sichuan, been caught in the vicinity of Chengdu, have Gansu and Shaanxi, together with even fewer appeared in Chengdu bird market (Wang 2004). presumed non-breeding records from Yunnan All other records are from Thailand and presumed (southern China) and northern Thailand. It is an to be either wintering birds or passage migrants extremely poorly known species classified as (BirdLife International 2001). Vulnerable because it is inferred to have a small, declining population as a result of destruction of Observations temperate forest within its breeding area as well On the morning of 2 May 2011, we visited Sichuan as habitat loss in its likely wintering areas (BirdLife University to observe and photograph migrating International 2001, 2011). birds in a small patch of wood and shrubbery in Since its description in 1891, there have only the south-eastern corner of the campus. At about been nine records from China, including some 09h20 a bird abruptly flew into the shrubbery, uncertain ones. These records comprise one each which we quickly located and identified as a male from Ganshu and Shaanxi, two from Yunnan, Blackthroat Luscinia obscura. We continued to presumed to be during migration, and four from observe it until about 17h20 when we left and Sichuan, all detailed in BirdLife International obtained what we believe to be the first images of (2001), and a recent sight record of a male and this species in the wild (Plates 1 & 2). -
![The Song Structure of the Siberian Blue Robin Luscinia [Larvivora] Cyane and a Comparison with Related Species](https://docslib.b-cdn.net/cover/7826/the-song-structure-of-the-siberian-blue-robin-luscinia-larvivora-cyane-and-a-comparison-with-related-species-897826.webp)
The Song Structure of the Siberian Blue Robin Luscinia [Larvivora] Cyane and a Comparison with Related Species
Ornithol Sci 16: 71 – 77 (2017) ORIGINAL ARTICLE The song structure of the Siberian Blue Robin Luscinia [Larvivora] cyane and a comparison with related species Vladimir IVANITSKII1,#, Alexandra IVLIEVA2, Sergey GASHKOV3 and Irina MAROVA1 1 Lomonosov Moscow State University, Biology, 119899 Moscow Leninskie Gory, Moscow, Moscow 119899, Russian Federation 2 M.F. Vladimirskii Moscow Regional Research Clinical Institute, Moscow, Moscow, Russian Federation 3 Tomsk State University-Zoology Museum, Tomsk, Tomsk, Russian Federation ORNITHOLOGICAL Abstract We studied the song syntax of the Siberian Blue Robin Luscinia cyane, a small insectivorous passerine of the taiga forests of Siberia and the Far East. Males SCIENCE have repertoires of 7 to 14 (mean 10.9±2.3) song types. A single song typically con- © The Ornithological Society sists of a short trill comprised of from three to six identical syllables, each of two to of Japan 2017 three notes; sometimes the trill is preceded by a short single note. The most complex songs contain as many as five or six different trills and single notes. The song of the Siberian Blue Robin most closely resembles that of the Indian Blue Robin L. brunnea. The individual repertoires of Siberian Blue Robin, Common Nightingale L. megarhynchos and Thrush Nightingale L. luscinia contain groups of mutually associ- ated song types that are sung usually one after another. The Siberian Blue Robin and the Common Nightingale perform them in a varying sequence, while Thrush Nightin- gale predominantly uses a fixed sequence of song types. The distinctions between the song syntax of Larvivora spp. and Luscinia spp. are discussed. -

Biodiversity Profile of Afghanistan
NEPA Biodiversity Profile of Afghanistan An Output of the National Capacity Needs Self-Assessment for Global Environment Management (NCSA) for Afghanistan June 2008 United Nations Environment Programme Post-Conflict and Disaster Management Branch First published in Kabul in 2008 by the United Nations Environment Programme. Copyright © 2008, United Nations Environment Programme. This publication may be reproduced in whole or in part and in any form for educational or non-profit purposes without special permission from the copyright holder, provided acknowledgement of the source is made. UNEP would appreciate receiving a copy of any publication that uses this publication as a source. No use of this publication may be made for resale or for any other commercial purpose whatsoever without prior permission in writing from the United Nations Environment Programme. United Nations Environment Programme Darulaman Kabul, Afghanistan Tel: +93 (0)799 382 571 E-mail: [email protected] Web: http://www.unep.org DISCLAIMER The contents of this volume do not necessarily reflect the views of UNEP, or contributory organizations. The designations employed and the presentations do not imply the expressions of any opinion whatsoever on the part of UNEP or contributory organizations concerning the legal status of any country, territory, city or area or its authority, or concerning the delimitation of its frontiers or boundaries. Unless otherwise credited, all the photos in this publication have been taken by the UNEP staff. Design and Layout: Rachel Dolores -

Federal Register/Vol. 85, No. 74/Thursday, April 16, 2020/Notices
21262 Federal Register / Vol. 85, No. 74 / Thursday, April 16, 2020 / Notices acquisition were not included in the 5275 Leesburg Pike, Falls Church, VA Comment (1): We received one calculation for TDC, the TDC limit would not 22041–3803; (703) 358–2376. comment from the Western Energy have exceeded amongst other items. SUPPLEMENTARY INFORMATION: Alliance, which requested that we Contact: Robert E. Mulderig, Deputy include European starling (Sturnus Assistant Secretary, Office of Public Housing What is the purpose of this notice? vulgaris) and house sparrow (Passer Investments, Office of Public and Indian Housing, Department of Housing and Urban The purpose of this notice is to domesticus) on the list of bird species Development, 451 Seventh Street SW, Room provide the public an updated list of not protected by the MBTA. 4130, Washington, DC 20410, telephone (202) ‘‘all nonnative, human-introduced bird Response: The draft list of nonnative, 402–4780. species to which the Migratory Bird human-introduced species was [FR Doc. 2020–08052 Filed 4–15–20; 8:45 am]‘ Treaty Act (16 U.S.C. 703 et seq.) does restricted to species belonging to biological families of migratory birds BILLING CODE 4210–67–P not apply,’’ as described in the MBTRA of 2004 (Division E, Title I, Sec. 143 of covered under any of the migratory bird the Consolidated Appropriations Act, treaties with Great Britain (for Canada), Mexico, Russia, or Japan. We excluded DEPARTMENT OF THE INTERIOR 2005; Pub. L. 108–447). The MBTRA states that ‘‘[a]s necessary, the Secretary species not occurring in biological Fish and Wildlife Service may update and publish the list of families included in the treaties from species exempted from protection of the the draft list. -

A Checklist of the Birds of Goa, India
BAIDYA & BHAGAT: Goa checklist 1 A checklist of the birds of Goa, India Pronoy Baidya & Mandar Bhagat Baidya, P., & Bhagat, M., 2018. A checklist of the birds of Goa, India. Indian BIRDS 14 (1): 1–31. Pronoy Baidya, TB-03, Center for Ecological Sciences, Indian Institute of Science, Bengaluru 560012, Karnataka, India. And, Foundation for Environment Research and Conservation, C/o 407, III-A, Susheela Seawinds, Alto-Vaddem, Vasco-da-Gama 403802, Goa, India. E-mail: [email protected] [Corresponding author] [PB] Mandar Bhagat, ‘Madhumangal’, New Vaddem,Vasco-da-Gama 403802, Goa, India. E-mail: [email protected] [MB] Manuscript received on 15 November 2017. We dedicate this paper to Heinz Lainer, for his commitment to Goa’s Ornithology. Abstract An updated checklist of the birds of Goa, India, is presented below based upon a collation of supporting information from museum specimens, photographs, audio recordings of calls, and sight records with sufficient field notes. Goa has 473 species of birds of which 11 are endemic to the Western Ghats, 19 fall under various categories of the IUCN Red List of Threatened Species, and 48 are listed in Schedule I Part (III) of The Indian Wild Life (Protection) Act, 1972. 451 species have been accepted into the checklist based on specimens in various museums or on photographs, while 22 have been accepted based on sight record. A secondary list of unconfirmed records is also discussed in detail. Introduction that is about 125 km long. The southern portion of these ghats, Goa, India’s smallest state, sandwiched between the Arabian within Goa, juts out towards the Arabian Sea, at Cabo de Rama, Sea in the west and the Western Ghats in the east, is home to and then curves inland. -
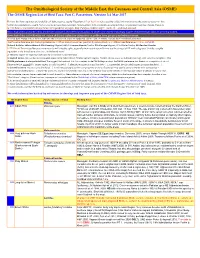
OSME List V3.4 Passerines-2
The Ornithological Society of the Middle East, the Caucasus and Central Asia (OSME) The OSME Region List of Bird Taxa: Part C, Passerines. Version 3.4 Mar 2017 For taxa that have unproven and probably unlikely presence, see the Hypothetical List. Red font indicates either added information since the previous version or that further documentation is sought. Not all synonyms have been examined. Serial numbers (SN) are merely an administrative conveninence and may change. Please do not cite them as row numbers in any formal correspondence or papers. Key: Compass cardinals (eg N = north, SE = southeast) are used. Rows shaded thus and with yellow text denote summaries of problem taxon groups in which some closely-related taxa may be of indeterminate status or are being studied. Rows shaded thus and with white text contain additional explanatory information on problem taxon groups as and when necessary. A broad dark orange line, as below, indicates the last taxon in a new or suggested species split, or where sspp are best considered separately. The Passerine Reference List (including References for Hypothetical passerines [see Part E] and explanations of Abbreviated References) follows at Part D. Notes↓ & Status abbreviations→ BM=Breeding Migrant, SB/SV=Summer Breeder/Visitor, PM=Passage Migrant, WV=Winter Visitor, RB=Resident Breeder 1. PT=Parent Taxon (used because many records will antedate splits, especially from recent research) – we use the concept of PT with a degree of latitude, roughly equivalent to the formal term sensu lato , ‘in the broad sense’. 2. The term 'report' or ‘reported’ indicates the occurrence is unconfirmed. -

EUROPEAN BIRDS of CONSERVATION CONCERN Populations, Trends and National Responsibilities
EUROPEAN BIRDS OF CONSERVATION CONCERN Populations, trends and national responsibilities COMPILED BY ANNA STANEVA AND IAN BURFIELD WITH SPONSORSHIP FROM CONTENTS Introduction 4 86 ITALY References 9 89 KOSOVO ALBANIA 10 92 LATVIA ANDORRA 14 95 LIECHTENSTEIN ARMENIA 16 97 LITHUANIA AUSTRIA 19 100 LUXEMBOURG AZERBAIJAN 22 102 MACEDONIA BELARUS 26 105 MALTA BELGIUM 29 107 MOLDOVA BOSNIA AND HERZEGOVINA 32 110 MONTENEGRO BULGARIA 35 113 NETHERLANDS CROATIA 39 116 NORWAY CYPRUS 42 119 POLAND CZECH REPUBLIC 45 122 PORTUGAL DENMARK 48 125 ROMANIA ESTONIA 51 128 RUSSIA BirdLife Europe and Central Asia is a partnership of 48 national conservation organisations and a leader in bird conservation. Our unique local to global FAROE ISLANDS DENMARK 54 132 SERBIA approach enables us to deliver high impact and long term conservation for the beneit of nature and people. BirdLife Europe and Central Asia is one of FINLAND 56 135 SLOVAKIA the six regional secretariats that compose BirdLife International. Based in Brus- sels, it supports the European and Central Asian Partnership and is present FRANCE 60 138 SLOVENIA in 47 countries including all EU Member States. With more than 4,100 staf in Europe, two million members and tens of thousands of skilled volunteers, GEORGIA 64 141 SPAIN BirdLife Europe and Central Asia, together with its national partners, owns or manages more than 6,000 nature sites totaling 320,000 hectares. GERMANY 67 145 SWEDEN GIBRALTAR UNITED KINGDOM 71 148 SWITZERLAND GREECE 72 151 TURKEY GREENLAND DENMARK 76 155 UKRAINE HUNGARY 78 159 UNITED KINGDOM ICELAND 81 162 European population sizes and trends STICHTING BIRDLIFE EUROPE GRATEFULLY ACKNOWLEDGES FINANCIAL SUPPORT FROM THE EUROPEAN COMMISSION. -

Eradication of the Mongoose Is Crucial for the Conservation of Three Endemic Bird Species in Yambaru, Okinawa Island, Japan
Biol Invasions https://doi.org/10.1007/s10530-021-02503-w (0123456789().,-volV)(0123456789().,-volV) ORIGINAL PAPER Eradication of the mongoose is crucial for the conservation of three endemic bird species in Yambaru, Okinawa Island, Japan Tsutomu Yagihashi . Shin-Ichi Seki . Tomoki Nakaya . Katsushi Nakata . Nobuhiko Kotaka Received: 4 September 2020 / Accepted: 20 March 2021 Ó The Author(s) 2021 Abstract Okinawa Island, Japan, is a globally results showed that the distribution areas of these bird important biodiversity hotspot. Three endemic bird species have been recovering since the 2007 within the species, Okinawa rail (Hypotaenidia okinawae), Oki- small Indian mongoose (Urva auropunctata) con- nawa woodpecker (Dendrocopos noguchii), and Oki- trolled area. The GAMM results showed that these nawa robin (Larvivora namiyei), are found only in the bird species were abundant in areas with fewer small Yambaru region of the northern part of Okinawa Indian mongooses and larger areas of hardwood Island. In order to conserve endemic species, it is forests. Thus, the mongoose had a negative impact important to determine the effect of alien species on not only on the flightless rails but also on the endemic species. We conducted playback surveys four woodpeckers and the robins. In recent years, most of times every three years from 2007 to 2016 to evaluate the old-growth forests have been designated as the recent distribution of these three forest-dwelling protected forests, and large-scale logging is no longer bird species during the breeding season. Then, the taking place in Yambaru. Eradication of the mongoose association between the numbers of detections of these is particularly important for the conservation of these three species with the invasive mongoose density and three endemic bird species.