Technical Tips for Obtaining Reliable DNA Identification of Historic Human Remains
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Washington University Record, April 1, 1993
Washington University School of Medicine Digital Commons@Becker Washington University Record Washington University Publications 4-1-1993 Washington University Record, April 1, 1993 Follow this and additional works at: http://digitalcommons.wustl.edu/record Recommended Citation "Washington University Record, April 1, 1993" (1993). Washington University Record. Book 615. http://digitalcommons.wustl.edu/record/615 This Article is brought to you for free and open access by the Washington University Publications at Digital Commons@Becker. It has been accepted for inclusion in Washington University Record by an authorized administrator of Digital Commons@Becker. For more information, please contact [email protected]. WASHINGTON UNIVERSITY IN ST. LOUIS iecord Vol. 17 No. 25 April 1,1993 Architecture class teaches universal design, sensitivity Architecture students recently tried experiencing the world from the perspective of people with disabilities. For a few hours, students maneuvered in wheelchairs, wore blindfolds over their eyes or plugs in their ears to try to get a sense of the needs of people with physi- cal impairments. Some students also spent an entire day in wheelchairs. One took the bus to the St. Louis Galleria, another tried to go out to dinner. For Michele Lewis, a graduate stu- dent in the course, just being at home was difficult. "I even had trouble getting into my building, and there are people living there who are in wheelchairs," she said. "The hallways are too skinny, I couldn't reach my toothbrush and the stove is too high. How are you supposed to know when your water is boiling when you can't see it?" This empathic experience is part of a course taught by Mary Ann Lazarus, vice president at Hellmuth, Obata, and Kassabaum (HOK), an international Sophomore Becky Sladky signs up to be a host for April Welcome, an expanded student recruitment program. -

The Polymerase Chain Reaction (PCR): the Second Generation of DNA Analysis Methods Takes the Stand, 9 Santa Clara High Tech
Santa Clara High Technology Law Journal Volume 9 | Issue 1 Article 8 January 1993 The olP ymerase Chain Reaction (PCR): The Second Generation of DNA Analysis Methods Takes the Stand Kamrin T. MacKnight Follow this and additional works at: http://digitalcommons.law.scu.edu/chtlj Part of the Law Commons Recommended Citation Kamrin T. MacKnight, The Polymerase Chain Reaction (PCR): The Second Generation of DNA Analysis Methods Takes the Stand, 9 Santa Clara High Tech. L.J. 287 (1993). Available at: http://digitalcommons.law.scu.edu/chtlj/vol9/iss1/8 This Comment is brought to you for free and open access by the Journals at Santa Clara Law Digital Commons. It has been accepted for inclusion in Santa Clara High Technology Law Journal by an authorized administrator of Santa Clara Law Digital Commons. For more information, please contact [email protected]. THE POLYMERASE CHAIN REACTION (PCR): THE SECOND GENERATION OF DNA ANALYSIS METHODS TAKES THE STAND Kamrin T. MacKnightt TABLE OF CONTENTS INTRODUCTION ........................................... 288 BASIC GENETICS AND DNA REPLICATION ................. 289 FORENSIC DNA ANALYSIS ................................ 292 Direct Sequencing ....................................... 293 Restriction FragmentLength Polymorphism (RFLP) ...... 294 Introduction .......................................... 294 Technology ........................................... 296 Polymerase Chain Reaction (PCR) ....................... 300 H istory ............................................... 300 Technology .......................................... -

Mittelalterliche Migrationen Als Gegenstand Der "Genetic History"
Jörg Feuchter Mittelalterliche Migrationen als Gegenstand der ‚Genetic History‘ Zusammenfassung Genetic History untersucht historische Fragen mit der Quelle DNA. Migrationen sind ihr Hauptgegenstand, woraus sich eine große Bedeutung für die Mediävistik ergibt. Doch bis vor kurzem waren Historiker nicht beteiligt. Der Beitrag gibt eine Definition der neuen Dis- ziplin (), untersucht die Rolle des Konzeptes Migration in der Populationsgenetik () und beschreibt den Stand der Migrationsforschung in der Mediävistik (). Anschließend gibt er einen kurzen Überblick über die von der Genetic History untersuchten Migrationsräu- me (), betrachtet exemplarisch zunächst eine Studie zur angelsächsischen Migration nach Britannien () und dann ein aktuelles Projekt, in dem erstmals ein Mediävist die Leitung innehat (). Der Beitrag endet mit einigen allgemeinen Beobachtungen und Postulaten (). Keywords: Genetik; DNA; Mittelalter; Angelsachsen; Langobarden; Völkerwanderung. Genetic History is the study of historical questions with DNA as a source. Migrations are its main subject. It is thus very relevant to Medieval Studies. Yet until recently historians have not been involved. The contribution provides a definition of the new discipline (), explores the role of migration as a concept in Population Genetics () and describes the state of migration studies in Medieval History (). It then sets out for an overview of medieval migratory areas studied by Genetic History () and takes an exemplary look first () at a study on Anglo-Saxon migration to Britain and later () at a current project where for the first time a medieval historian has taken the lead. The contribution ends () with some general observations and stipulations. Keywords: Genetics; DNA; Middle Ages; Anglo-Saxons; Lombards; Migration Period. Danksagung: Der Verfasser dankt Prof. Dr. Veronika Lipphardt (University College Frei- burg) und der von ihr von bis geleiteten Nachwuchsgruppe Twentieth Century Felix Wiedemann, Kerstin P. -

AMPK Against NASH Non-Alcoholic Steatohepatitis (NASH) Is Activation Were Elevated in LAKO Mice the Most Severe Form of Non-Alcoholic on NASH-Inducing Diets
RESEARCH HIGHLIGHTS collected from people representing published the first report of Journal Club all major global populations. mtDNA mutations causing a The most important discovery of disease — Leber’s hereditary optic mtDNA IN THE CROSSROADS OF their work was that the highest neuropathy. As disease- causing EVOLUTION AND DISEASE degree of mtDNA variation was mutations are amenable to natural found among Africans, attesting to selection, how could one consider Mitochondrial DNA (mtDNA) their anti quity. The other finding the mtDNA to be merely a neutral sequences are currently studied was that although mtDNA from marker? It was this contradiction mainly by three disciplines: across the globe contained African that drew me into studying molecular evolution (including types, Africans had many unique mitochondrial biology and to anthropology), functional mtDNA types. discovering that even common genomics and analyses of genetic These two findings, in addition population variants of mtDNA are disorders. Strangely, these discip- to other data, prompted Cann et al. both subjected to natural selection lines can use the relatively short mtDNA (1987) to propose that the origin and alter the tendency to develop sequence of mtDNA in opposing has been of all current human populations genetic disorders. manners, by attributing to it either considered (at least the maternal lineage) is Dan Mishmar Department of Life Sciences, lack of function, as is the case an excellent likely African. Many criticized in (some) molecular evolution this study, especially because Ben- Gurion University of the Negev, marker for Be’er- Sheva, Israel. studies, or conversely, functional their African samples originated e- mail: [email protected] mainly from Afro-Americans relevance in the aetiology of tracing ancient The author declares no competing interests genetic disorders. -
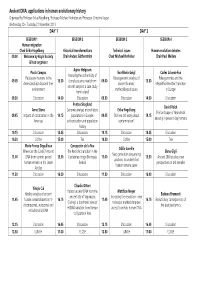
Final Programme
Ancient DNA: applications in human evolutionary history Organised by Professor Erika Hagelberg, Professor Michael Hofreiter and Professor Christine Keyser Wednesday 20 - Thursday 21 November 2013 DAY 1 DAY 2 SESSION 1 SESSION 2 SESSION 3 SESSION 4 Human migration Chair Erika Hagelberg Historical transformations Technical issues Human evolution debates 09.00 Welcome by Royal Society Chair Anders Götherström Chair Michael Hofreiter Chair Paul Mellars & lead organiser Agnar Helgason Paula Campos Eva-Maria Geigl Carles Lalueza-Fox Assessing the authenticity of Pleistocene humans in the Palaeogenomic analysis of Paleogenomics and the 09.05 13.30 clonal sequence reads from 09.00 13.30 Americans/East Asia and their ancient humans: Mesolithic-Neolithic transition ancient samples: a case study environment methodological issues in Europe from Iceland 09.30 Discussion 14.00 Discussion 09.30 Discussion 14.00 Discussion Pontus Skoglund David Reich Anne Stone Genomic analysis of prehistoric Erika Hagelberg The landscape of Neandertal 09.45 Impacts of colonization in the 14.15 populations in Europe: 09.45 Shall we still worry about 14.15 ancestry in present-day humans Americas authentication and population contamination? history 10.15 Discussion 14.45 Discussion 10.15 Discussion 14.45 Discussion 10.30 Coffee 15.00 Tea 10.30 Coffee 15.00 Tea Marie-France Deguilloux Concepción de la Rua Odille Loreille Where are the Caribs? Ancient The Neolithic transition in the Elena Gigli Next generation sequencing 11.00 DNA from ceramic period 15.30 Cantabrian fringe -

Profile of Alec J. Jeffreys
Profile of Alec J. Jeffreys s one of the great contributors to modern genetics, Sir Alec Jeffreys was born with curiosity in his genes as the son and Agrandson of prolific inventors. Jeffreys displayed an insatiable quest for knowl- edge, and his father fostered his son’s budding scientific interests with gifts of a microscope and chemistry set, the latter of which produced one of Jeffreys’ most memorable scientific ventures. ‘‘Those were back in the happy days of chemis- try,’’ Jeffreys points out, ‘‘where you could go down to your local pharmacist and get virtually everything you wanted.’’ The end result of that chemistry experiment was the detonation of his aunt’s apple tree and a set of scars that Jeffreys still bears to- day. ‘‘You learn science very fast that way,’’ he says, ‘‘but it was quite fun.’’ Jeffreys’ scientific curiosity only in- creased after the apple tree incident, and years later it would lead to one of the Alec J. Jeffreys most widely used applications in genetics: DNA fingerprinting. Among other uses, the DNA fingerprinting technique has freys go there, so Jeffreys entered the turn led to another groundbreaking find- helped solve numerous criminal investi- university in 1968. He was at first intent ing about the composition of eukaryotic gations, settle countless paternity dis- on pursuing a degree in biochemistry DNA: introns (3). ‘‘I was 27 at the time, putes, and spark a resurgence of interest but soon decided to alter his course. so still a real rookie, with the ability to in the forensic sciences. That achieve- ‘‘This was no criticism of the way it was detect single-copy DNA by Southern blot ment alone is worthy of merit, contrib- taught,’’ he says of biochemistry at Ox- hybridization and a nice paper on introns uting to Jeffreys’ receiving three high ford, ‘‘but rather a reflection of the fac- under my belt, which is, yeah, not a bad distinctions in 2005: the Albert Lasker ulty’s research, which leaned heavily to start,’’ he says. -

Recent Human Evolution
A common ancestor at 500 ka? H. heid. in Europe and Africa LCA of Nea. and sapiens? The origin of our species (H. sapiens) TWO origins to explain: 1.The shared (species) features 2. Non-shared (regional/racial) features Origins of modern human behaviour? Complex technology “Houses Networking” Symbolic burials Art Models of modern human origins 1984 1970 1987: Mitochondrial Eve hits the headlines! Mitochondrial DNA and human evolution Nature 325, 31-36 Rebecca L. Cann, Mark Stoneking & Allan C. Wilson (1987) African female ancestor ~200ka The pendulum starts swinging! 2000 1995 1995 1984 1970 The African record H. sapiens: fossils suggest an African origin for the modern pattern ~ 150-200ka? Age ka ~260 ~150? ~160? ~195? >130 Tim White Lieberman Pinnacle Point Howiesonspoort Microliths Klasies Twin Rivers Enakpune Mumba ya Muto Shellfishing Grotta Moscerini Ochre Kapthurin Twin Rivers Klasies Qafzeh Blombos 300 ka 200 150 100 50 0 Taforalt Shell beads Skhul Enakpune Blombos ya Muto Early H. sapiens fossils Omo Kibish Herto “Modern” anatomy and behaviour have deep roots in Africa… ~60 ka: Modern Humans start to leave Africa… ? Out of Africa Mitochondrial DNA Out of Africa Y-chromosome DNA The evolution of regionality (“race”) DNA benetton Natural selection Bottlenecks + Founder Effects Sexual/cultural selection We are all the same (species), but we all look different (individuals, ♀/♂, regions, “races”). Species Individuals (Homo sapiens) large brain Natural Selection body shape high round skull Sexual Selection skin colour small face hair -

Unique Features of Human Skin
Center for Academic Research & Training in Anthropogeny (CARTA) Unique Features of Human Skin Public Symposium ! Friday, October 16, 2015 Chairs: Nina Jablonski, Pennsylvania State University Pascal Gagneux, UC San Diego This CARTA symposium is sponsored by: The G. Harold and Leila Y. Mathers Charitable Foundation ABSTRACTS Skin: A Window into the Evolution of the Human Super-Organism Richard Gallo, UC San Diego All life must establish an external barrier that simultaneously interacts with and defends itself from the outside world. Skin is our most important barrier, and human skin has many functions that are in common with other animals but also unique to our species. General skin functions include sensing danger, regulation of internal temperature, maintaining fluid balance and mounting visual sexual displays. Human life also depends on the capacity of our skin to protect us from specific environmental dangers and pathogens unique to us. The central teachings of the medical specialty of Dermatology have been that health is defined by the capacity of skin to perform all these functions while constantly resisting entry of microbial pathogens. However, a recent revolution in understanding skin biology has revealed that normal skin functions are performed not only by cells of human origin, but also by specialized microbes that have co-evolved with us to live in a mutually beneficial relationship. This presentation will provide an overview of the multiple cell types, both human and microbial, that comprise the human skin super-organism. Understanding this relationship changes how we should think about evolution, gene transfer and the impact of current hygiene and antibiotic therapies. -

Patterns of Human Diversity, Within and Among Continents, Inferred from Biallelic DNA Polymorphisms
Letter Patterns of Human Diversity, within and among Continents, Inferred from Biallelic DNA Polymorphisms Chiara Romualdi,1,2,7 David Balding,3,8 Ivane S. Nasidze,4 Gregory Risch,5 Myles Robichaux,5 Stephen T. Sherry,5 Mark Stoneking,4 Mark A. Batzer,5,6 and Guido Barbujani1,9 1 Department of Biology, University of Ferrara, Ferrara I-44100, Italy; 2 Department of Statistics, University of Padua, Padua I-35121, Italy; 3 Department of Applied Statistics, University of Reading, Reading RG6 6FN, United Kingdom; 4 Max Planck Institute for Evolutionary Anthropology, Leipzig, D-04103 Germany; 5 Department of Pathology, Stanley S. Scott Cancer Center, Louisiana State University Health Sciences Center, New Orleans, Louisiana 70112, USA; 6 Department of Biological Sciences, Biological Computation and Visualization Center, Louisiana State University, Baton Rouge, Louisiana 70803, USA Previous studies have reported that about 85% of human diversity at Short Tandem Repeat (STR) and Restriction Fragment Length Polymorphism (RFLP) autosomal loci is due to differences between individuals of the same population, whereas differences among continental groups account for only 10% of the overall genetic variance. These findings conflict with popular notions of distinct and relatively homogeneous human races, and may also call into question the apparent usefulness of ethnic classification in, for example, medical diagnostics. Here, we present new data on 21 Alu insertions in 32 populations. We analyze these data along with three other large, globally dispersed data sets consisting of apparently neutral biallelic nuclear markers, as well as with a -globin data set possibly subject to selection. We confirm the previous results for the autosomal data, and find a higher diversity among continents for Y-chromosome loci. -

The New Science of Human Evolution - Newsweek Technology -
The New Science of Human Evolution - Newsweek Technology - ... http://www.msnbc.msn.com/id/17542627/site/newsweek/print/1/dis... MSNBC.com The New Science of Human Evolution The new science of the brain and DNA is rewriting the history of human origins. B y Sh ar on Be gl ey Newsweek March 19, 2007 issue - Unlike teeth and skulls and other bones, hair is no match for the pitiless ravages of weather, geologic upheaval and time. So although skulls from millions of years ago testify to the increase in brain size as one species of human ancestor evolved into the next, and although the architecture of spine and hips shows when our ancestors first stood erect, the fossil record is silent on when they fully lost their body hair and replaced it with clothing. Which makes it fortunate that Mark Stoneking thought of lice. Head lice live in the hair on the head. But body lice, a larger variety, are misnamed: they live in clothing. Head lice, as a species, go back millions of years, while body lice are a more recent arrival. Stoneking, an evolutionary anthropologist, had a hunch that he could calculate when body lice evolved from head lice by comparing the two varieties' DNA, which accumulates changes at a regular rate. (It's like calculating how long it took a typist to produce a document if you know he makes six typos per minute.) That fork in the louse's family tree, he and colleagues at Germany's Max Planck Institute for Evolutionary Anthropology concluded, occurred no more than 114,000 years ago. -

The DNA Fingerprinting Story Mark a Jobling
Jobling Investigative Genetics 2013, 4:20 http://www.investigativegenetics.com/content/4/1/20 OPINION Open Access Curiosity in the genes: the DNA fingerprinting story Mark A Jobling It is unusual for a scientific field to be associated with a The first DNA fingerprinting application was in par- single individual, but in the case of the subject of the the- entage testing [6]; normally it is the father who is in matic series now being launched in Investigative Genetics, doubt, but this unusual and challenging case was a ma- this is surely so; Alec Jeffreys (Figure 1) is DNA finger- ternity test, with paternal DNA unavailable. British nurse printing. Having invented the method, he coined the per- Christiana Sarbah’s 13-year-old son Andrew was denied fect name for it - how different things might have been if re-entry to the United Kingdom after a visit to Ghana, it had been called the tandem-repeat-based identification the immigration authorities suspecting that he was not technique (or something similarly dull). He realized its her child. Given three undisputed children for comparison, potential and immediately applied, developed and refined it was possible to reconstruct the absent father’sDNA it. He then followed his nose to unravel the mystery of fingerprint, and to strongly support the claimed maternity the madly mutable minisatellites that make up DNA over alternative relationships such as aunt-nephew - some- fingerprints, and eventually to understand the engines of thing that was not achieved with traditional protein poly- genome variability that reside in recombination hotspots. morphisms such as blood groups. -

Past Human Migrations in East Asia: Matching Archaeology, Linguistics
16 A genetic perspective on the origins and dispersal of the Austronesians Mitochondrial DNA variation from Madagascar to Easter Island Erika Hagelberg, Murray Cox, Wulf Schiefenhövel, and Ian Frame Introduction This chapter describes the study of one genetic system, mitochondrial DNA (mtDNA), within the Austronesian-speaking world, and discusses whether the patterns of genetic variation at this genetic locus are consistent with the principal models of the settlement and linguistic diversity in this vast geographical region. Mitochondria are the seat of cellular metabolism and have their own complement of DNA. MtDNA is a tiny proportion of the genetic makeup of a human being, and has both advantages and disadvantages in population studies. It is inherited maternally, in contrast with the nuclear chromosomal DNA inherited from both parents. (Note: the mitochondria of the spermatozoon are eliminated from the egg soon after fertilization, so offspring do not inherit paternal mtDNA, although there are exceptions to this rule, as shown by Schwartz and Vissing 2002.) The maternal mode of inheritance permits the study of female lineages through time. Despite the small size of the mitochondrial genome, each mitochondrion has many thousands of copies of mtDNA, and there are many mitochondria in each cell. The abundance of mtDNA in cells has been exploited in anthropological research on ancient, rare or degraded biological samples, including archaeological bones, old hair samples, or dried blood (Hagelberg 1994). MtDNA accumulates mutations at a relatively fast rate, and so is useful for the investigation of recent evolutionary events. These features have made mtDNA an excellent marker for geneticists interested in human evolutionary history (see Wallace 1995 for review).