Genomic and Transcriptomic Analysis of Carbohydrate Utilization by Paenibacillus Sp
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplementary Information
doi: 10.1038/nature06269 SUPPLEMENTARY INFORMATION METAGENOMIC AND FUNCTIONAL ANALYSIS OF HINDGUT MICROBIOTA OF A WOOD FEEDING HIGHER TERMITE TABLE OF CONTENTS MATERIALS AND METHODS 2 • Glycoside hydrolase catalytic domains and carbohydrate binding modules used in searches that are not represented by Pfam HMMs 5 SUPPLEMENTARY TABLES • Table S1. Non-parametric diversity estimators 8 • Table S2. Estimates of gross community structure based on sequence composition binning, and conserved single copy gene phylogenies 8 • Table S3. Summary of numbers glycosyl hydrolases (GHs) and carbon-binding modules (CBMs) discovered in the P3 luminal microbiota 9 • Table S4. Summary of glycosyl hydrolases, their binning information, and activity screening results 13 • Table S5. Comparison of abundance of glycosyl hydrolases in different single organism genomes and metagenome datasets 17 • Table S6. Comparison of abundance of glycosyl hydrolases in different single organism genomes (continued) 20 • Table S7. Phylogenetic characterization of the termite gut metagenome sequence dataset, based on compositional phylogenetic analysis 23 • Table S8. Counts of genes classified to COGs corresponding to different hydrogenase families 24 • Table S9. Fe-only hydrogenases (COG4624, large subunit, C-terminal domain) identified in the P3 luminal microbiota. 25 • Table S10. Gene clusters overrepresented in termite P3 luminal microbiota versus soil, ocean and human gut metagenome datasets. 29 • Table S11. Operational taxonomic unit (OTU) representatives of 16S rRNA sequences obtained from the P3 luminal fluid of Nasutitermes spp. 30 SUPPLEMENTARY FIGURES • Fig. S1. Phylogenetic identification of termite host species 38 • Fig. S2. Accumulation curves of 16S rRNA genes obtained from the P3 luminal microbiota 39 • Fig. S3. Phylogenetic diversity of P3 luminal microbiota within the phylum Spirocheates 40 • Fig. -

United States Patent (19) 11 Patent Number: 5,981,835 Austin-Phillips Et Al
USOO598.1835A United States Patent (19) 11 Patent Number: 5,981,835 Austin-Phillips et al. (45) Date of Patent: Nov. 9, 1999 54) TRANSGENIC PLANTS AS AN Brown and Atanassov (1985), Role of genetic background in ALTERNATIVE SOURCE OF Somatic embryogenesis in Medicago. Plant Cell Tissue LIGNOCELLULOSC-DEGRADING Organ Culture 4:107-114. ENZYMES Carrer et al. (1993), Kanamycin resistance as a Selectable marker for plastid transformation in tobacco. Mol. Gen. 75 Inventors: Sandra Austin-Phillips; Richard R. Genet. 241:49-56. Burgess, both of Madison; Thomas L. Castillo et al. (1994), Rapid production of fertile transgenic German, Hollandale; Thomas plants of Rye. Bio/Technology 12:1366–1371. Ziegelhoffer, Madison, all of Wis. Comai et al. (1990), Novel and useful properties of a chimeric plant promoter combining CaMV 35S and MAS 73 Assignee: Wisconsin Alumni Research elements. Plant Mol. Biol. 15:373-381. Foundation, Madison, Wis. Coughlan, M.P. (1988), Staining Techniques for the Detec tion of the Individual Components of Cellulolytic Enzyme 21 Appl. No.: 08/883,495 Systems. Methods in Enzymology 160:135-144. de Castro Silva Filho et al. (1996), Mitochondrial and 22 Filed: Jun. 26, 1997 chloroplast targeting Sequences in tandem modify protein import specificity in plant organelles. Plant Mol. Biol. Related U.S. Application Data 30:769-78O. 60 Provisional application No. 60/028,718, Oct. 17, 1996. Divne et al. (1994), The three-dimensional crystal structure 51 Int. Cl. ............................. C12N 15/82; C12N 5/04; of the catalytic core of cellobiohydrolase I from Tricho AO1H 5/00 derma reesei. Science 265:524-528. -

WO 2015/130881 Al 3 September 2015 (03.09.2015) P O P CT
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2015/130881 Al 3 September 2015 (03.09.2015) P O P CT (51) International Patent Classification: (72) Inventors: NAGY, Kevin D.; 106 Steven Lane, Wilming C12P 19/16 (2006.01) C12P 19/12 (2006.01) ton, Delaware 19808 (US). HAGO, Erwin Columbus; CUP 19/18 (2006.01) C12P 19/08 (2006.01) 2901 6th Street Sw Apt. 24, Cedar Rapids, Iowa 52404 CUP 19/04 (2006.01) (US). SHETTY, Jayarama K.; 4806 Braxton Place, Pleasonton, California 94566 (US). HENNESSEY, Susan (21) International Application Number: Marie; 32 Truman Lane, Avondale, Pennsylvania 193 11 PCT/US20 15/0 17644 (US). DICOSIMO, Robert; 1607 Masters Way, Chadds (22) International Filing Date: Ford, Pennsylvania 193 17-9720 (US). HUA, Ling; 126 26 February 2015 (26.02.2015) Hockessin Drive, Hockessin, Delaware 19707 (US). RAMIREZ, Rodrigo; Rua Alfredo Ribeiro Nogueira, 280. (25) Filing Language: English Casa08, Campinas, 13092-480 Sao Paulo (BR). TANG, Publication Language: English Zhongmei; Room 602, Building 100, Lane 1100, Gudai Road, Shanghai 201 102 (CN). YU, Zheyong; Room 501, Priority Data: Building 17, Lane 150, Guangyue Road, Hongkou District, 61/945,233 27 February 2014 (27.02.2014) US Shanghai 200000 (CN). 61/945,241 27 February 2014 (27.02.2014) US 62/004,300 29 May 2014 (29.05.2014) us (74) Agent: CHESIRE, Dennis; E. I. du Pont de Nemours and 62/004,305 29 May 2014 (29.05.2014) us Company, Legal Patent Records Center, Chestnut Run 62/004,308 29 May 2014 (29.05.2014) us Plaza 721/2640, 974 Centre Road, PO Box 291 5 Wilming 62/004,290 29 May 2014 (29.05.2014) us ton, Delaware 19805 (US). -

(12) United States Patent (10) Patent No.: US 6,468,955 B1 Smets Et Al
USOO6468955B1 (12) United States Patent (10) Patent No.: US 6,468,955 B1 Smets et al. (45) Date of Patent: Oct. 22, 2002 (54) LAUNDRY DETERGENT AND/OR FABRIC FOREIGN PATENT DOCUMENTS CARE COMPOSITIONS COMPRISINGA MODIFIED ENZYME WO WO 94/07998 4/1994 ............ C12N/9/42 WO WO 94/29460 12/1994 ... C12N/15/62 (75) Inventors: Johan Smets, Lubbeek (BE); Jean-Luc WO WO 97/07203 2/1997 C12N/11/12 Philippe Bettiol, Brussels (BE); WO WO 97/24421 7/1997 WO WO-97/24421 * 7/1997 ........... C11D/3/386 Stanton Lane Boyer, Fairfield, OH WO WO-97/28243 * 8/1997 ..... ... C11D/3/386 (US); Alfred Busch, Londerzeel (BE) WO WO 97/28256 8/1997 ..... ... C12N/9/00 WO WO 97/301.48 8/1997 ..... ... C12N/9/96 (73) Assignee: The Proctor & Gamble Company, WO WO 98/OO500 1/1998 ..... ... C11D/3/386 Cincinnati, OH (US) WO WO 98/16633 4/1998 ..... ... C12N/9/26 (*) Notice: Subject to any disclaimer, the term of this WO WO 98/284.11 7/1998 ............ C12N/9/42 patent is extended or adjusted under 35 U.S.C. 154(b) by 0 days. OTHER PUBLICATIONS (21) Appl. No.: 09/674,478 Characterization of a Double Cellulose-Binding Domain No (22) PCT Filed: Apr. 30, 1999 date. (86) PCT No.: PCT/US99/09453 * cited by examiner S371 (c)(1), (2), (4) Date: Nov. 1, 2000 Primary Examiner Mark Kopec (87) PCT Pub. No.: WO99/57252 ASSistant Examiner-Eisa Elhilo (74) Attorney, Agent, or Firm-Frank Taffy; C. Brant Cook; PCT Pub. Date: Nov. 11, 1999 K. -

(12) United States Patent (10) Patent No.: US 9,689,046 B2 Mayall Et Al
USOO9689046B2 (12) United States Patent (10) Patent No.: US 9,689,046 B2 Mayall et al. (45) Date of Patent: Jun. 27, 2017 (54) SYSTEM AND METHODS FOR THE FOREIGN PATENT DOCUMENTS DETECTION OF MULTIPLE CHEMICAL WO O125472 A1 4/2001 COMPOUNDS WO O169245 A2 9, 2001 (71) Applicants: Robert Matthew Mayall, Calgary (CA); Emily Candice Hicks, Calgary OTHER PUBLICATIONS (CA); Margaret Mary-Flora Bebeselea, A. et al., “Electrochemical Degradation and Determina Renaud-Young, Calgary (CA); David tion of 4-Nitrophenol Using Multiple Pulsed Amperometry at Christopher Lloyd, Calgary (CA); Lisa Graphite Based Electrodes', Chem. Bull. “Politehnica” Univ. Kara Oberding, Calgary (CA); Iain (Timisoara), vol. 53(67), 1-2, 2008. Fraser Scotney George, Calgary (CA) Ben-Yoav. H. et al., “A whole cell electrochemical biosensor for water genotoxicity bio-detection”. Electrochimica Acta, 2009, 54(25), 6113-6118. (72) Inventors: Robert Matthew Mayall, Calgary Biran, I. et al., “On-line monitoring of gene expression'. Microbi (CA); Emily Candice Hicks, Calgary ology (Reading, England), 1999, 145 (Pt 8), 2129-2133. (CA); Margaret Mary-Flora Da Silva, P.S. et al., “Electrochemical Behavior of Hydroquinone Renaud-Young, Calgary (CA); David and Catechol at a Silsesquioxane-Modified Carbon Paste Elec trode'. J. Braz. Chem. Soc., vol. 24, No. 4, 695-699, 2013. Christopher Lloyd, Calgary (CA); Lisa Enache, T. A. & Oliveira-Brett, A. M., "Phenol and Para-Substituted Kara Oberding, Calgary (CA); Iain Phenols Electrochemical Oxidation Pathways”, Journal of Fraser Scotney George, Calgary (CA) Electroanalytical Chemistry, 2011, 1-35. Etesami, M. et al., “Electrooxidation of hydroquinone on simply prepared Au-Pt bimetallic nanoparticles'. Science China, Chem (73) Assignee: FREDSENSE TECHNOLOGIES istry, vol. -

Generate Metabolic Map Poster
Authors: Pallavi Subhraveti Ron Caspi Quang Ong Peter D Karp An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_900114035Cyc: Amycolatopsis sacchari DSM 44468 Cellular Overview Connections between pathways are omitted for legibility. -

12) United States Patent (10
US007635572B2 (12) UnitedO States Patent (10) Patent No.: US 7,635,572 B2 Zhou et al. (45) Date of Patent: Dec. 22, 2009 (54) METHODS FOR CONDUCTING ASSAYS FOR 5,506,121 A 4/1996 Skerra et al. ENZYME ACTIVITY ON PROTEIN 5,510,270 A 4/1996 Fodor et al. MICROARRAYS 5,512,492 A 4/1996 Herron et al. 5,516,635 A 5/1996 Ekins et al. (75) Inventors: Fang X. Zhou, New Haven, CT (US); 5,532,128 A 7/1996 Eggers Barry Schweitzer, Cheshire, CT (US) 5,538,897 A 7/1996 Yates, III et al. s s 5,541,070 A 7/1996 Kauvar (73) Assignee: Life Technologies Corporation, .. S.E. al Carlsbad, CA (US) 5,585,069 A 12/1996 Zanzucchi et al. 5,585,639 A 12/1996 Dorsel et al. (*) Notice: Subject to any disclaimer, the term of this 5,593,838 A 1/1997 Zanzucchi et al. patent is extended or adjusted under 35 5,605,662 A 2f1997 Heller et al. U.S.C. 154(b) by 0 days. 5,620,850 A 4/1997 Bamdad et al. 5,624,711 A 4/1997 Sundberg et al. (21) Appl. No.: 10/865,431 5,627,369 A 5/1997 Vestal et al. 5,629,213 A 5/1997 Kornguth et al. (22) Filed: Jun. 9, 2004 (Continued) (65) Prior Publication Data FOREIGN PATENT DOCUMENTS US 2005/O118665 A1 Jun. 2, 2005 EP 596421 10, 1993 EP 0619321 12/1994 (51) Int. Cl. EP O664452 7, 1995 CI2O 1/50 (2006.01) EP O818467 1, 1998 (52) U.S. -

(12) Patent Application Publication (10) Pub. No.: US 2011/0165635 A1 Copenhaver Et Al
US 2011 O165635A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2011/0165635 A1 Copenhaver et al. (43) Pub. Date: Jul. 7, 2011 (54) METHODS AND MATERALS FOR Publication Classification PROCESSINGA FEEDSTOCK (51) Int. Cl. CI2P I 7/04 (2006.01) (75) Inventors: Gregory P. Copenhaver, Chapel CI2P I/00 (2006.01) Hill, NC (US); Daphne Preuss, CI2P 7/04 (2006.01) Chicago, IL (US); Jennifer Mach, CI2P 7/16 (2006.01) Chicago, IL (US) CI2P 7/06 (2006.01) CI2P 5/00 (2006.01) CI2P 5/02 (2006.01) (73) Assignee: CHROMATIN, INC., Chicago, IL CI2P3/00 (2006.01) (US) CI2P I/02 (2006.01) CI2N 5/10 (2006.01) (21) Appl. No.: 12/989,038 CI2N L/15 (2006.01) CI2N I/3 (2006.01) (52) U.S. Cl. ........... 435/126; 435/41; 435/157; 435/160; (22) PCT Fled: Apr. 21, 2009 435/161; 435/166; 435/167; 435/168; 435/171; 435/419,435/254.11: 435/257.2 (86) PCT NO.: PCT/US2O09/041260 (57) ABSTRACT S371 (c)(1), The present disclosure relates generally to methods for pro (2), (4) Date: Mar. 11, 2011 cessing a feedstock. Specifically, methods are provided for processing a feedstock by mixing the feedstock with an addi tive organism that comprises one or more transgenes coding Related U.S. Application Data for one or more enzymes. The expressed enzymes may be (60) Provisional application No. 61/046,705, filed on Apr. capable of breaking down cellulosic and lignocellulosic 21, 2008. materials and converting them to a biofuel. -
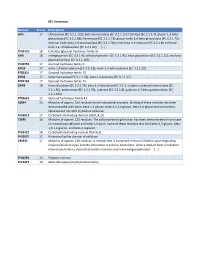
Beta-Mannosidase (EC 3.2.1.25)
M1 Consensus Domain # runs Description GH5 17 chitosanase (EC 3.2.1.132); beta-mannosidase (EC 3.2.1.25); Cellulase (EC 3.2.1.4); glucan 1,3-beta- glucosidase (EC 3.2.1.58); licheninase (EC 3.2.1.73); glucan endo-1,6-beta-glucosidase (EC 3.2.1.75); mannan endo-beta-1,4-mannosidase (EC 3.2.1.78); endo-beta-1,4-xylanase (EC 3.2.1.8); cellulose beta-1,4-cellobiosidase (EC 3.2.1.91); [...] PF00150 18 Cellulase (glycosyl hydrolase family 5) GH9 18 endoglucanase (EC 3.2.1.4); cellobiohydrolase (EC 3.2.1.91); beta-glucosidase (EC 3.2.1.21); exo-beta- glucosaminidase (EC 3.2.1.165) PF00759 17 Glycosyl hydrolase family 9 GH10 17 endo-1,4-beta-xylanase (EC 3.2.1.8); endo-1,3-beta-xylanase (EC 3.2.1.32) PF00331 17 Glycosyl hydrolase family 10 GH26 17 beta-mannanase (EC 3.2.1.78); beta-1,3-xylanase (EC 3.2.1.32) PF02156 17 Glycosyl hydrolase family 26 GH43 18 beta-xylosidase (EC 3.2.1.37); beta-1,3-xylosidase (EC 3.2.1.-); alpha-L-arabinofuranosidase (EC 3.2.1.55); arabinanase (EC 3.2.1.99); xylanase (EC 3.2.1.8); galactan 1,3-beta-galactosidase (EC 3.2.1.145) PF04616 17 Glycosyl hydrolases family 43 CBM4 16 Modules of approx. 150 residues found in bacterial enzymes. Binding of these modules has been demonstrated with xylan, beta-1,3-glucan, beta-1,3-1,4-glucan, beta-1,6-glucan and amorphous cellulose but not with crystalline cellulose. -

Molecular Functions, Catalysis and Reguation Beilstein Enzymology
Molecular Functions, Catalysis and Reguation Beilstein Enzymology Symposium 2019 10 – 12 September 2019 Hotel Jagdschloss Niederwald Rüdesheim, Germany The Beilstein-Institut and Open Science The non-profit Beilstein-Institut is one of the most respected organizations in the communication and dissemination of high-quality information in chemistry. Since 1951, when the foundation was established by the Max Planck Society, we have been fulfilling our mission to support the scientific community by providing high-quality information that is essential for research. Our role has evolved over the years: from the production of the Beilstein Handbook and Database, to being one of the first open access journal publishers in chemistry, to host of interdisciplinary symposia and supporter of open data initiatives. We believe that free access to scientific research results, giving everyone in the world an equal chance to read and reuse experimental findings and data, is the best way to advance science. Open Science is a new approach to scientific research. It is based on cooperation and uses new ways to disseminate information and broaden knowledge through digital technologies and new collaborative tools. It aims to make the primary outputs of publicly funded research results – publications (open access) and the research data (open data) – publicly accessible in digital format with no or minimal restriction. he !eilstein"Institut supports open science and makes the results of its pro#ects freely available to the scientific community as open access publications. his is an essential contribution to the foundation’s mission to advance the chemical and related sciences. %ll #ournal articles& conference proceedings and videos are open access to allow the worldwide& unhindered sharing and e'change of ideas. -

(12) Patent Application Publication (10) Pub. No.: US 2012/0266329 A1 Mathur Et Al
US 2012026.6329A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2012/0266329 A1 Mathur et al. (43) Pub. Date: Oct. 18, 2012 (54) NUCLEICACIDS AND PROTEINS AND CI2N 9/10 (2006.01) METHODS FOR MAKING AND USING THEMI CI2N 9/24 (2006.01) CI2N 9/02 (2006.01) (75) Inventors: Eric J. Mathur, Carlsbad, CA CI2N 9/06 (2006.01) (US); Cathy Chang, San Marcos, CI2P 2L/02 (2006.01) CA (US) CI2O I/04 (2006.01) CI2N 9/96 (2006.01) (73) Assignee: BP Corporation North America CI2N 5/82 (2006.01) Inc., Houston, TX (US) CI2N 15/53 (2006.01) CI2N IS/54 (2006.01) CI2N 15/57 2006.O1 (22) Filed: Feb. 20, 2012 CI2N IS/60 308: Related U.S. Application Data EN f :08: (62) Division of application No. 1 1/817,403, filed on May AOIH 5/00 (2006.01) 7, 2008, now Pat. No. 8,119,385, filed as application AOIH 5/10 (2006.01) No. PCT/US2006/007642 on Mar. 3, 2006. C07K I4/00 (2006.01) CI2N IS/II (2006.01) (60) Provisional application No. 60/658,984, filed on Mar. AOIH I/06 (2006.01) 4, 2005. CI2N 15/63 (2006.01) Publication Classification (52) U.S. Cl. ................... 800/293; 435/320.1; 435/252.3: 435/325; 435/254.11: 435/254.2:435/348; (51) Int. Cl. 435/419; 435/195; 435/196; 435/198: 435/233; CI2N 15/52 (2006.01) 435/201:435/232; 435/208; 435/227; 435/193; CI2N 15/85 (2006.01) 435/200; 435/189: 435/191: 435/69.1; 435/34; CI2N 5/86 (2006.01) 435/188:536/23.2; 435/468; 800/298; 800/320; CI2N 15/867 (2006.01) 800/317.2: 800/317.4: 800/320.3: 800/306; CI2N 5/864 (2006.01) 800/312 800/320.2: 800/317.3; 800/322; CI2N 5/8 (2006.01) 800/320.1; 530/350, 536/23.1: 800/278; 800/294 CI2N I/2 (2006.01) CI2N 5/10 (2006.01) (57) ABSTRACT CI2N L/15 (2006.01) CI2N I/19 (2006.01) The invention provides polypeptides, including enzymes, CI2N 9/14 (2006.01) structural proteins and binding proteins, polynucleotides CI2N 9/16 (2006.01) encoding these polypeptides, and methods of making and CI2N 9/20 (2006.01) using these polynucleotides and polypeptides. -

L-Helical Catalytic Domains in Glycoside Hydrolase Families 49, 55 and 87: Domain Architecture, Modelling and Assignment of Catalytic Residues
FEBS 26656 FEBS Letters 530 (2002) 225^232 L-Helical catalytic domains in glycoside hydrolase families 49, 55 and 87: domain architecture, modelling and assignment of catalytic residues Daniel J. Rigdena;Ã, Octa¤vio L. Francoa;b aEmbrapa Genetic Resources and Biotechnology, Cenargen/Embrapa, S.A.I.N. Parque Rural, Final W5, Asa Norte, 70770-900 Bras|¤lia DF, Brazil bUniversidade Cato¤lica de Bras|¤lia, Bras|¤lia DF, Brazil Received 24 July 2002; revised 19 September 2002; accepted 19 September 2002 First published online 3 October 2002 Edited by Robert B. Russell hydrolases (GHs; EC 3.2.1.-). These have been classi¢ed Abstract X-ray crystallography and bioinformatics studies re- veal a tendency for the right-handed L-helix domain architecture according to similarity in sequence (and structure, where to be associated with carbohydrate binding proteins. Here we available) into 88 families in the CAZY database [7,8] now demonstrate the presence of catalytic L-helix domains in glyco- accessible online [9]. Grouping in this way enables the deter- side hydrolase (GH) families 49, 55 and 87 and provide evidence mination of important characteristics of all members of a for their sharing a common evolutionary ancestor with two family based on experimental evidence obtained for one. Typ- structurally characterized GH families, numbers 28 and 82. ically interest focuses on the identity of the two acidic cata- This domain assignment helps assign catalytic residues to each lytic residues, nearly universally involved in cleavage of the family. Further analysis of domain architecture reveals the as- glycoside bond [10], and whether the mechanism is of the sociation of carbohydrate binding modules with catalytic GH inverting or retaining type.