Hierarchical Models – Motivation James-Stein Inference • Suppose X ∼ N(Θ, 1) – X Is Admissible (Not Dominated) for Estimating Θ with Squared Error Loss
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Assessing Fairness with Unlabeled Data and Bayesian Inference
Can I Trust My Fairness Metric? Assessing Fairness with Unlabeled Data and Bayesian Inference Disi Ji1 Padhraic Smyth1 Mark Steyvers2 1Department of Computer Science 2Department of Cognitive Sciences University of California, Irvine [email protected] [email protected] [email protected] Abstract We investigate the problem of reliably assessing group fairness when labeled examples are few but unlabeled examples are plentiful. We propose a general Bayesian framework that can augment labeled data with unlabeled data to produce more accurate and lower-variance estimates compared to methods based on labeled data alone. Our approach estimates calibrated scores for unlabeled examples in each group using a hierarchical latent variable model conditioned on labeled examples. This in turn allows for inference of posterior distributions with associated notions of uncertainty for a variety of group fairness metrics. We demonstrate that our approach leads to significant and consistent reductions in estimation error across multiple well-known fairness datasets, sensitive attributes, and predictive models. The results show the benefits of using both unlabeled data and Bayesian inference in terms of assessing whether a prediction model is fair or not. 1 Introduction Machine learning models are increasingly used to make important decisions about individuals. At the same time it has become apparent that these models are susceptible to producing systematically biased decisions with respect to sensitive attributes such as gender, ethnicity, and age [Angwin et al., 2017, Berk et al., 2018, Corbett-Davies and Goel, 2018, Chen et al., 2019, Beutel et al., 2019]. This has led to a significant amount of recent work in machine learning addressing these issues, including research on both (i) definitions of fairness in a machine learning context (e.g., Dwork et al. -

Bayesian Inference Chapter 4: Regression and Hierarchical Models
Bayesian Inference Chapter 4: Regression and Hierarchical Models Conchi Aus´ınand Mike Wiper Department of Statistics Universidad Carlos III de Madrid Master in Business Administration and Quantitative Methods Master in Mathematical Engineering Conchi Aus´ınand Mike Wiper Regression and hierarchical models Masters Programmes 1 / 35 Objective AFM Smith Dennis Lindley We analyze the Bayesian approach to fitting normal and generalized linear models and introduce the Bayesian hierarchical modeling approach. Also, we study the modeling and forecasting of time series. Conchi Aus´ınand Mike Wiper Regression and hierarchical models Masters Programmes 2 / 35 Contents 1 Normal linear models 1.1. ANOVA model 1.2. Simple linear regression model 2 Generalized linear models 3 Hierarchical models 4 Dynamic models Conchi Aus´ınand Mike Wiper Regression and hierarchical models Masters Programmes 3 / 35 Normal linear models A normal linear model is of the following form: y = Xθ + ; 0 where y = (y1;:::; yn) is the observed data, X is a known n × k matrix, called 0 the design matrix, θ = (θ1; : : : ; θk ) is the parameter set and follows a multivariate normal distribution. Usually, it is assumed that: 1 ∼ N 0 ; I : k φ k A simple example of normal linear model is the simple linear regression model T 1 1 ::: 1 where X = and θ = (α; β)T . x1 x2 ::: xn Conchi Aus´ınand Mike Wiper Regression and hierarchical models Masters Programmes 4 / 35 Normal linear models Consider a normal linear model, y = Xθ + . A conjugate prior distribution is a normal-gamma distribution: -

The Exponential Family 1 Definition
The Exponential Family David M. Blei Columbia University November 9, 2016 The exponential family is a class of densities (Brown, 1986). It encompasses many familiar forms of likelihoods, such as the Gaussian, Poisson, multinomial, and Bernoulli. It also encompasses their conjugate priors, such as the Gamma, Dirichlet, and beta. 1 Definition A probability density in the exponential family has this form p.x / h.x/ exp >t.x/ a./ ; (1) j D f g where is the natural parameter; t.x/ are sufficient statistics; h.x/ is the “base measure;” a./ is the log normalizer. Examples of exponential family distributions include Gaussian, gamma, Poisson, Bernoulli, multinomial, Markov models. Examples of distributions that are not in this family include student-t, mixtures, and hidden Markov models. (We are considering these families as distributions of data. The latent variables are implicitly marginalized out.) The statistic t.x/ is called sufficient because the probability as a function of only depends on x through t.x/. The exponential family has fundamental connections to the world of graphical models (Wainwright and Jordan, 2008). For our purposes, we’ll use exponential 1 families as components in directed graphical models, e.g., in the mixtures of Gaussians. The log normalizer ensures that the density integrates to 1, Z a./ log h.x/ exp >t.x/ d.x/ (2) D f g This is the negative logarithm of the normalizing constant. The function h.x/ can be a source of confusion. One way to interpret h.x/ is the (unnormalized) distribution of x when 0. It might involve statistics of x that D are not in t.x/, i.e., that do not vary with the natural parameter. -

Machine Learning Conjugate Priors and Monte Carlo Methods
Hierarchical Bayes for Non-IID Data Conjugate Priors Monte Carlo Methods CPSC 540: Machine Learning Conjugate Priors and Monte Carlo Methods Mark Schmidt University of British Columbia Winter 2016 Hierarchical Bayes for Non-IID Data Conjugate Priors Monte Carlo Methods Admin Nothing exciting? We discussed empirical Bayes, where you optimize prior using marginal likelihood, Z argmax p(xjα; β) = argmax p(xjθ)p(θjα; β)dθ: α,β α,β θ Can be used to optimize λj, polynomial degree, RBF σi, polynomial vs. RBF, etc. We also considered hierarchical Bayes, where you put a prior on the prior, p(xjα; β)p(α; βjγ) p(α; βjx; γ) = : p(xjγ) But is the hyper-prior really needed? Hierarchical Bayes for Non-IID Data Conjugate Priors Monte Carlo Methods Last Time: Bayesian Statistics In Bayesian statistics we work with posterior over parameters, p(xjθ)p(θjα; β) p(θjx; α; β) = : p(xjα; β) We also considered hierarchical Bayes, where you put a prior on the prior, p(xjα; β)p(α; βjγ) p(α; βjx; γ) = : p(xjγ) But is the hyper-prior really needed? Hierarchical Bayes for Non-IID Data Conjugate Priors Monte Carlo Methods Last Time: Bayesian Statistics In Bayesian statistics we work with posterior over parameters, p(xjθ)p(θjα; β) p(θjx; α; β) = : p(xjα; β) We discussed empirical Bayes, where you optimize prior using marginal likelihood, Z argmax p(xjα; β) = argmax p(xjθ)p(θjα; β)dθ: α,β α,β θ Can be used to optimize λj, polynomial degree, RBF σi, polynomial vs. -

Practical Statistics for Particle Physics Lecture 1 AEPS2018, Quy Nhon, Vietnam
Practical Statistics for Particle Physics Lecture 1 AEPS2018, Quy Nhon, Vietnam Roger Barlow The University of Huddersfield August 2018 Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 1 / 34 Lecture 1: The Basics 1 Probability What is it? Frequentist Probability Conditional Probability and Bayes' Theorem Bayesian Probability 2 Probability distributions and their properties Expectation Values Binomial, Poisson and Gaussian 3 Hypothesis testing Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 2 / 34 Question: What is Probability? Typical exam question Q1 Explain what is meant by the Probability PA of an event A [1] Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 3 / 34 Four possible answers PA is number obeying certain mathematical rules. PA is a property of A that determines how often A happens For N trials in which A occurs NA times, PA is the limit of NA=N for large N PA is my belief that A will happen, measurable by seeing what odds I will accept in a bet. Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 4 / 34 Mathematical Kolmogorov Axioms: For all A ⊂ S PA ≥ 0 PS = 1 P(A[B) = PA + PB if A \ B = ϕ and A; B ⊂ S From these simple axioms a complete and complicated structure can be − ≤ erected. E.g. show PA = 1 PA, and show PA 1.... But!!! This says nothing about what PA actually means. Kolmogorov had frequentist probability in mind, but these axioms apply to any definition. Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 5 / 34 Classical or Real probability Evolved during the 18th-19th century Developed (Pascal, Laplace and others) to serve the gambling industry. -
3.3 Bayes' Formula
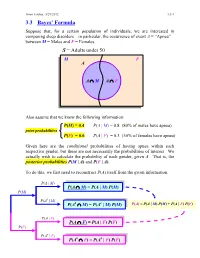
Ismor Fischer, 5/29/2012 3.3-1 3.3 Bayes’ Formula Suppose that, for a certain population of individuals, we are interested in comparing sleep disorders – in particular, the occurrence of event A = “Apnea” – between M = Males and F = Females. S = Adults under 50 M F A A ∩ M A ∩ F Also assume that we know the following information: P(M) = 0.4 P(A | M) = 0.8 (80% of males have apnea) prior probabilities P(F) = 0.6 P(A | F) = 0.3 (30% of females have apnea) Given here are the conditional probabilities of having apnea within each respective gender, but these are not necessarily the probabilities of interest. We actually wish to calculate the probability of each gender, given A. That is, the posterior probabilities P(M | A) and P(F | A). To do this, we first need to reconstruct P(A) itself from the given information. P(A | M) P(A ∩ M) = P(A | M) P(M) P(M) P(Ac | M) c c P(A ∩ M) = P(A | M) P(M) P(A) = P(A | M) P(M) + P(A | F) P(F) P(A | F) P(A ∩ F) = P(A | F) P(F) P(F) P(Ac | F) c c P(A ∩ F) = P(A | F) P(F) Ismor Fischer, 5/29/2012 3.3-2 So, given A… P(M ∩ A) P(A | M) P(M) P(M | A) = P(A) = P(A | M) P(M) + P(A | F) P(F) (0.8)(0.4) 0.32 = (0.8)(0.4) + (0.3)(0.6) = 0.50 = 0.64 and posterior P(F ∩ A) P(A | F) P(F) P(F | A) = = probabilities P(A) P(A | M) P(M) + P(A | F) P(F) (0.3)(0.6) 0.18 = (0.8)(0.4) + (0.3)(0.6) = 0.50 = 0.36 S Thus, the additional information that a M F randomly selected individual has apnea (an A event with probability 50% – why?) increases the likelihood of being male from a prior probability of 40% to a posterior probability 0.32 0.18 of 64%, and likewise, decreases the likelihood of being female from a prior probability of 60% to a posterior probability of 36%. -

Naïve Bayes Classifier
CS 60050 Machine Learning Naïve Bayes Classifier Some slides taken from course materials of Tan, Steinbach, Kumar Bayes Classifier ● A probabilistic framework for solving classification problems ● Approach for modeling probabilistic relationships between the attribute set and the class variable – May not be possible to certainly predict class label of a test record even if it has identical attributes to some training records – Reason: noisy data or presence of certain factors that are not included in the analysis Probability Basics ● P(A = a, C = c): joint probability that random variables A and C will take values a and c respectively ● P(A = a | C = c): conditional probability that A will take the value a, given that C has taken value c P(A,C) P(C | A) = P(A) P(A,C) P(A | C) = P(C) Bayes Theorem ● Bayes theorem: P(A | C)P(C) P(C | A) = P(A) ● P(C) known as the prior probability ● P(C | A) known as the posterior probability Example of Bayes Theorem ● Given: – A doctor knows that meningitis causes stiff neck 50% of the time – Prior probability of any patient having meningitis is 1/50,000 – Prior probability of any patient having stiff neck is 1/20 ● If a patient has stiff neck, what’s the probability he/she has meningitis? P(S | M )P(M ) 0.5×1/50000 P(M | S) = = = 0.0002 P(S) 1/ 20 Bayesian Classifiers ● Consider each attribute and class label as random variables ● Given a record with attributes (A1, A2,…,An) – Goal is to predict class C – Specifically, we want to find the value of C that maximizes P(C| A1, A2,…,An ) ● Can we estimate -

Polynomial Singular Value Decompositions of a Family of Source-Channel Models
Polynomial Singular Value Decompositions of a Family of Source-Channel Models The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation Makur, Anuran and Lizhong Zheng. "Polynomial Singular Value Decompositions of a Family of Source-Channel Models." IEEE Transactions on Information Theory 63, 12 (December 2017): 7716 - 7728. © 2017 IEEE As Published http://dx.doi.org/10.1109/tit.2017.2760626 Publisher Institute of Electrical and Electronics Engineers (IEEE) Version Author's final manuscript Citable link https://hdl.handle.net/1721.1/131019 Terms of Use Creative Commons Attribution-Noncommercial-Share Alike Detailed Terms http://creativecommons.org/licenses/by-nc-sa/4.0/ IEEE TRANSACTIONS ON INFORMATION THEORY 1 Polynomial Singular Value Decompositions of a Family of Source-Channel Models Anuran Makur, Student Member, IEEE, and Lizhong Zheng, Fellow, IEEE Abstract—In this paper, we show that the conditional expec- interested in a particular subclass of one-parameter exponential tation operators corresponding to a family of source-channel families known as natural exponential families with quadratic models, defined by natural exponential families with quadratic variance functions (NEFQVF). So, we define natural exponen- variance functions and their conjugate priors, have orthonor- mal polynomials as singular vectors. These models include the tial families next. Gaussian channel with Gaussian source, the Poisson channel Definition 1 (Natural Exponential Family). Given a measur- with gamma source, and the binomial channel with beta source. To derive the singular vectors of these models, we prove and able space (Y; B(Y)) with a σ-finite measure µ, where Y ⊆ R employ the equivalent condition that their conditional moments and B(Y) denotes the Borel σ-algebra on Y, the parametrized are strictly degree preserving polynomials. -

Numerical Physics with Probabilities: the Monte Carlo Method and Bayesian Statistics Part I for Assignment 2
Numerical Physics with Probabilities: The Monte Carlo Method and Bayesian Statistics Part I for Assignment 2 Department of Physics, University of Surrey module: Energy, Entropy and Numerical Physics (PHY2063) 1 Numerical Physics part of Energy, Entropy and Numerical Physics This numerical physics course is part of the second-year Energy, Entropy and Numerical Physics mod- ule. It is online at the EENP module on SurreyLearn. See there for assignments, deadlines etc. The course is about numerically solving ODEs (ordinary differential equations) and PDEs (partial differential equations), and introducing the (large) part of numerical physics where probabilities are used. This assignment is on numerical physics of probabilities, and looks at the Monte Carlo (MC) method, and at the Bayesian statistics approach to data analysis. It covers MC and Bayesian statistics, in that order. MC is a widely used numerical technique, it is used, amongst other things, for modelling many random processes. MC is used in fields from statistical physics, to nuclear and particle physics. Bayesian statistics is a powerful data analysis method, and is used everywhere from particle physics to spam-email filters. Data analysis is fundamental to science. For example, analysis of the data from the Large Hadron Collider was required to extract a most probable value for the mass of the Higgs boson, together with an estimate of the region of masses where the scientists think the mass is. This region is typically expressed as a range of mass values where the they think the true mass lies with high (e.g., 95%) probability. Many of you will be analysing data (physics data, commercial data, etc) for your PTY or RY, or future careers1 . -

The Bayesian Approach to Statistics
THE BAYESIAN APPROACH TO STATISTICS ANTHONY O’HAGAN INTRODUCTION the true nature of scientific reasoning. The fi- nal section addresses various features of modern By far the most widely taught and used statisti- Bayesian methods that provide some explanation for the rapid increase in their adoption since the cal methods in practice are those of the frequen- 1980s. tist school. The ideas of frequentist inference, as set out in Chapter 5 of this book, rest on the frequency definition of probability (Chapter 2), BAYESIAN INFERENCE and were developed in the first half of the 20th century. This chapter concerns a radically differ- We first present the basic procedures of Bayesian ent approach to statistics, the Bayesian approach, inference. which depends instead on the subjective defini- tion of probability (Chapter 3). In some respects, Bayesian methods are older than frequentist ones, Bayes’s Theorem and the Nature of Learning having been the basis of very early statistical rea- Bayesian inference is a process of learning soning as far back as the 18th century. Bayesian from data. To give substance to this statement, statistics as it is now understood, however, dates we need to identify who is doing the learning and back to the 1950s, with subsequent development what they are learning about. in the second half of the 20th century. Over that time, the Bayesian approach has steadily gained Terms and Notation ground, and is now recognized as a legitimate al- ternative to the frequentist approach. The person doing the learning is an individual This chapter is organized into three sections. -

The Bayesian Lasso
The Bayesian Lasso Trevor Park and George Casella y University of Florida, Gainesville, Florida, USA Summary. The Lasso estimate for linear regression parameters can be interpreted as a Bayesian posterior mode estimate when the priors on the regression parameters are indepen- dent double-exponential (Laplace) distributions. This posterior can also be accessed through a Gibbs sampler using conjugate normal priors for the regression parameters, with indepen- dent exponential hyperpriors on their variances. This leads to tractable full conditional distri- butions through a connection with the inverse Gaussian distribution. Although the Bayesian Lasso does not automatically perform variable selection, it does provide standard errors and Bayesian credible intervals that can guide variable selection. Moreover, the structure of the hierarchical model provides both Bayesian and likelihood methods for selecting the Lasso pa- rameter. The methods described here can also be extended to other Lasso-related estimation methods like bridge regression and robust variants. Keywords: Gibbs sampler, inverse Gaussian, linear regression, empirical Bayes, penalised regression, hierarchical models, scale mixture of normals 1. Introduction The Lasso of Tibshirani (1996) is a method for simultaneous shrinkage and model selection in regression problems. It is most commonly applied to the linear regression model y = µ1n + Xβ + ; where y is the n 1 vector of responses, µ is the overall mean, X is the n p matrix × T × of standardised regressors, β = (β1; : : : ; βp) is the vector of regression coefficients to be estimated, and is the n 1 vector of independent and identically distributed normal errors with mean 0 and unknown× variance σ2. The estimate of µ is taken as the average y¯ of the responses, and the Lasso estimate β minimises the sum of the squared residuals, subject to a given bound t on its L1 norm. -

Posterior Propriety and Admissibility of Hyperpriors in Normal
The Annals of Statistics 2005, Vol. 33, No. 2, 606–646 DOI: 10.1214/009053605000000075 c Institute of Mathematical Statistics, 2005 POSTERIOR PROPRIETY AND ADMISSIBILITY OF HYPERPRIORS IN NORMAL HIERARCHICAL MODELS1 By James O. Berger, William Strawderman and Dejun Tang Duke University and SAMSI, Rutgers University and Novartis Pharmaceuticals Hierarchical modeling is wonderful and here to stay, but hyper- parameter priors are often chosen in a casual fashion. Unfortunately, as the number of hyperparameters grows, the effects of casual choices can multiply, leading to considerably inferior performance. As an ex- treme, but not uncommon, example use of the wrong hyperparameter priors can even lead to impropriety of the posterior. For exchangeable hierarchical multivariate normal models, we first determine when a standard class of hierarchical priors results in proper or improper posteriors. We next determine which elements of this class lead to admissible estimators of the mean under quadratic loss; such considerations provide one useful guideline for choice among hierarchical priors. Finally, computational issues with the resulting posterior distributions are addressed. 1. Introduction. 1.1. The model and the problems. Consider the block multivariate nor- mal situation (sometimes called the “matrix of means problem”) specified by the following hierarchical Bayesian model: (1) X ∼ Np(θ, I), θ ∼ Np(B, Σπ), where X1 θ1 X2 θ2 Xp×1 = . , θp×1 = . , arXiv:math/0505605v1 [math.ST] 27 May 2005 . X θ m m Received February 2004; revised July 2004. 1Supported by NSF Grants DMS-98-02261 and DMS-01-03265. AMS 2000 subject classifications. Primary 62C15; secondary 62F15. Key words and phrases. Covariance matrix, quadratic loss, frequentist risk, posterior impropriety, objective priors, Markov chain Monte Carlo.