A Phylogenetic Framework for the Terns (Sternini) Inferred from Mtdna Sequences: Implications for Taxonomy and Plumage Evolution
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Conservation Problems on Tristan Da Cunha Byj
28 Oryx Conservation Problems on Tristan da Cunha ByJ. H. Flint The author spent two years, 1963-65, as schoolmaster on Tristan da Cunha, during which he spent four weeks on Nightingale Island. On the main island he found that bird stocks were being depleted and the islanders taking too many eggs and young; on Nightingale, however, where there are over two million pairs of great shearwaters, the harvest of these birds could be greater. Inaccessible Island, which like Nightingale, is without cats, dogs or rats, should be declared a wildlife sanctuary. Tl^HEN the first permanent settlers came to Tristan da Cunha in " the early years of the nineteenth century they found an island rich in bird and sea mammal life. "The mountains are covered with Albatross Mellahs Petrels Seahens, etc.," wrote Jonathan Lambert in 1811, and Midshipman Greene, who stayed on the island in 1816, recorded in his diary "Sea Elephants herding together in immense numbers." Today the picture is greatly changed. A century and a half of human habitation has drastically reduced the larger, edible species, and the accidental introduction of rats from a shipwreck in 1882 accelerated the birds' decline on the main island. Wood-cutting, grazing by domestic stock and, more recently, fumes from the volcano have destroyed much of the natural vegetation near the settlement, and two bird subspecies, a bunting and a flightless moorhen, have become extinct on the main island. Curiously, one is liable to see more birds on the day of arrival than in several weeks ashore. When I first saw Tristan from the decks of M.V. -

Order CHARADRIIFORMES: Waders, Gulls and Terns Suborder LARI
Text extracted from Gill B.J.; Bell, B.D.; Chambers, G.K.; Medway, D.G.; Palma, R.L.; Scofield, R.P.; Tennyson, A.J.D.; Worthy, T.H. 2010. Checklist of the birds of New Zealand, Norfolk and Macquarie Islands, and the Ross Dependency, Antarctica. 4th edition. Wellington, Te Papa Press and Ornithological Society of New Zealand. Pages 191, 223 & 227-228. Order CHARADRIIFORMES: Waders, Gulls and Terns The family sequence of Christidis & Boles (1994), who adopted that of Sibley et al. (1988) and Sibley & Monroe (1990), is followed here. Suborder LARI: Skuas, Gulls, Terns and Skimmers Condon (1975) and Checklist Committee (1990) recognised three subfamilies within the Laridae (Larinae, Sterninae and Megalopterinae) but this division has not been widely adopted. We follow Gochfeld & Burger (1996) in recognising gulls in one family (Laridae) and terns and noddies in another (Sternidae). The sequence of species for Stercorariidae and Laridae follows Peters (1934) and for Sternidae follows Bridge et al. (2005). Family LARIDAE Rafinesque: Gulls Laridia Rafinesque, 1815: Analyse de la Nature: 72 – Type genus Larus Linnaeus, 1758. Genus Larus Linnaeus Larus Linnaeus, 1758: Syst. Nat., 10th edition 1: 136 – Type species (by subsequent designation) Larus marinus Linnaeus. Gavia Boie, 1822: Isis von Oken, Heft 10: col. 563 – Type species (by subsequent designation) Larus ridibundus Linnaeus. Junior homonym of Gavia Moehring, 1758. Hydrocoleus Kaup, 1829: Skizz. Entw.-Gesch. Eur. Thierw.: 113 – Type species (by subsequent designation) Larus minutus Linnaeus. Chroicocephalus Eyton, 1836: Cat. Brit. Birds: 53 – Type species (by monotypy) Larus cucullatus Reichenbach = Larus pipixcan Wagler. Gelastes Bonaparte, 1853: Journ. für Ornith. -
Red-Footed Booby Helper at Great Frigatebird Nests
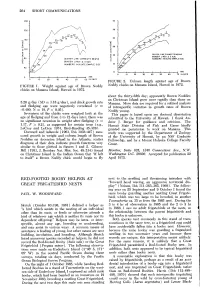
264 SHORT COMMUNICATIONS NECTS MEANS ECTS MEANS ICATE SAMPLE SIZE S.D. SAMPLE SIZE 70 IN DAYS FIGURE 2. Culmen length against age of Brown FIGURE 1. Weight against age of Brown Noddy Noddy chicks on Manana Island, Hawaii in 1972. chicks on Manana Island, Hawaii in 1972. about the thirty-fifth day; apparently Brown Noddies on Christmas Island grow more rapidly than those on 5.26 g/day (SD = 1.18 g/day), and chick growth rate Manana. More data are required for a refined analysis and fledging age were negatively correlated (r = of intraspecific variation in growth rates of Brown -0.490, N = 19, P < 0.05). Noddy young. Seventeen of the chicks were weighed both at the This paper is based upon my doctoral dissertation age of fledging and from 3 to 12 days later; there was submitted to the University of Hawaii. I thank An- no significant recession in weight after fledging (t = drew J. Berger for guidance and criticism. The 1.17, P > 0.2), as suggested for certain terns (e.g., Hawaii State Division of Fish and Game kindly LeCroy and LeCroy 1974, Bird-Banding 45:326). granted me permission to work on Manana. This Dorward and Ashmole (1963, Ibis 103b: 447) mea- study was supported by the Department of Zoology sured growth in weight and culmen length of Brown of the University of Hawaii, by an NSF Graduate Noddies on Ascension Island in the Atlantic; scatter Fellowship, and by a Mount Holyoke College Faculty diagrams of their data indicate growth functions very Grant. -

NOTORNIS 27 1980 LAKES of NORTH KAIPARA 3 for Observation Were Far from Suitable
NOTORNIS Journal of the Ornithological Society of New Zealand Volume 27 Part 1 March 1980 OFFICERS 1979 - 80 President - Mr. B. D. BELL, Wildlife Service, Dept. of Internal Affairs, Private Bag, Wellington Vice-president - Mr. M. L. FALCONER, 188 Miromiro Road, Normandale, Lower Hutt Editor - Mr. B. D. HEATHER, 10 Jocelyn Crescent, Silverstream Treasurer - Mr. H. W. M. HOGG, P.O. Box 3011, Dunedin Secretary - Mr R. S. SLACK, 31 Wyndham Road, Silverstream Council Members: Dr. BEN D. BELL, 45 Gurney Road, Belmont, Lower Hutt Mrs. B. BROWN, 39 Red Hill Road, Papakura Dr. P. C. BULL, 131A Waterloo Road, Lower Hutt Mr D. E. CROCKETT, 21 McMillan Avenue, Kamo, Whangarei Mr. F. C. KINSKY, 338 The Parade, Island Bay, Wellington 5 Mrs. S. M. REED, 4 Mamaku Street, Auckland 5 Mr. R. R. SUTTON, Lorneville, No. 4 R.D., Invercargill Conveners and Organisers: Rare Birds Committee (Acting): Mr. B. D. BELL Beach Patrol: Mr. C. R. VEITCH, Wildlife Service, Dept. of Internal Affairs, P.O. Box 2220, Auckland Card Committee: Mr. R. N. THOMAS, 25 Ravenswood Drive, Forest Hill, Auckland 10 Field Investigation Committee: Mr. B. D. BELL ~ibraria;: Miss A. J. GOODWIN, R.D. 1, Clevtdon Nest, Records: Mr. D. E. CROCKETT Recording (including material for Classified SU-arised Notes) : Mr. R. B. SIBSON, 26 Entrican Avenue, kemuera, Auckland Representative on Member Bodies' Committee of Royal Society of NX.: Mr. B. D. BELL Assistant Editor: Mr A. BLACKBURN, 10 Score Road, isb borne Editor of OSNZ ~ek:Mr'P. SAGAR, 38A Yardley St., Christchurch 4 .SUBSCRIPTIONS AND MEMBERSHIP Annual Subscription: Ordinary member $12; Husband & wife mem- bers $18; Junior'member (under 20) $9; Life mepber $240; Family member (one Notornis per household) ,bein other family of a member in. -

US Fish & Wildlife Service Seabird Conservation Plan—Pacific Region
U.S. Fish & Wildlife Service Seabird Conservation Plan Conservation Seabird Pacific Region U.S. Fish & Wildlife Service Seabird Conservation Plan—Pacific Region 120 0’0"E 140 0’0"E 160 0’0"E 180 0’0" 160 0’0"W 140 0’0"W 120 0’0"W 100 0’0"W RUSSIA CANADA 0’0"N 0’0"N 50 50 WA CHINA US Fish and Wildlife Service Pacific Region OR ID AN NV JAP CA H A 0’0"N I W 0’0"N 30 S A 30 N L I ort I Main Hawaiian Islands Commonwealth of the hwe A stern A (see inset below) Northern Mariana Islands Haw N aiian Isla D N nds S P a c i f i c Wake Atoll S ND ANA O c e a n LA RI IS Johnston Atoll MA Guam L I 0’0"N 0’0"N N 10 10 Kingman Reef E Palmyra Atoll I S 160 0’0"W 158 0’0"W 156 0’0"W L Howland Island Equator A M a i n H a w a i i a n I s l a n d s Baker Island Jarvis N P H O E N I X D IN D Island Kauai S 0’0"N ONE 0’0"N I S L A N D S 22 SI 22 A PAPUA NEW Niihau Oahu GUINEA Molokai Maui 0’0"S Lanai 0’0"S 10 AMERICAN P a c i f i c 10 Kahoolawe SAMOA O c e a n Hawaii 0’0"N 0’0"N 20 FIJI 20 AUSTRALIA 0 200 Miles 0 2,000 ES - OTS/FR Miles September 2003 160 0’0"W 158 0’0"W 156 0’0"W (800) 244-WILD http://www.fws.gov Information U.S. -

A Guide to the Birds of Barrow Island
A Guide to the Birds of Barrow Island Operated by Chevron Australia This document has been printed by a Sustainable Green Printer on stock that is certified carbon in joint venture with neutral and is Forestry Stewardship Council (FSC) mix certified, ensuring fibres are sourced from certified and well managed forests. The stock 55% recycled (30% pre consumer, 25% post- Cert no. L2/0011.2010 consumer) and has an ISO 14001 Environmental Certification. ISBN 978-0-9871120-1-9 Gorgon Project Osaka Gas | Tokyo Gas | Chubu Electric Power Chevron’s Policy on Working in Sensitive Areas Protecting the safety and health of people and the environment is a Chevron core value. About the Authors Therefore, we: • Strive to design our facilities and conduct our operations to avoid adverse impacts to human health and to operate in an environmentally sound, reliable and Dr Dorian Moro efficient manner. • Conduct our operations responsibly in all areas, including environments with sensitive Dorian Moro works for Chevron Australia as the Terrestrial Ecologist biological characteristics. in the Australasia Strategic Business Unit. His Bachelor of Science Chevron strives to avoid or reduce significant risks and impacts our projects and (Hons) studies at La Trobe University (Victoria), focused on small operations may pose to sensitive species, habitats and ecosystems. This means that we: mammal communities in coastal areas of Victoria. His PhD (University • Integrate biodiversity into our business decision-making and management through our of Western Australia) -

Sandwich Terns to Isla Rasa, Gulf of California, Mexico, with Our Observations of Single Individuals in 1986 and Again in 2008
NOTES SANDWICH TERNS ON ISLA RASA, GULF OF CALIFORNIA, MEXICO ENRIQUETA VELARDE, Instituto de Ciencias Marinas y Pesquerías, Universidad Veracruzana, Hidalgo 617, Col. Río Jamapa, Boca del Río, Veracruz, México, C.P. 94290; [email protected] MARISOL TORDESILLAS, Álvaro Obregón 549, Col. Poblado Alfredo V. Bonfi l, Cancún, Quintana Roo, México, C.P. 77560; [email protected] In North America the Sandwich Tern (Thalasseus sandvicensis) breeds locally along marine coasts and offshore islands primarily of the southeastern United States and Caribbean (AOU 1998). In these areas it commonly nests in dense colonies of the Royal Tern (T. maximus) and Laughing Gull (Leucophaeus atricilla; Shealer 1999). It winters along the coasts of the Atlantic Ocean and Gulf of Mexico from Florida to the West Indies, more rarely as far south as southern Brazil and Uruguay. It also winters on the Pacifi c coast, mainly from Oaxaca, Mexico, south to Panama (Howell and Webb 1995), occasionally to Colombia, Ecuador, and Peru (AOU 1998). As there are no breeding colonies on the Pacifi c coast, all birds wintering there are believed to represent migrants from Atlantic and Caribbean colonies (Collins 1997, Hilty and Brown 1986, Ridgely 1981, Ridgely and Greenfi eld 2001). Sandwich Terns have occasionally wandered as far north as eastern Canada (New Brunswick, Nova Scotia, and Newfoundland) and inland to Minnesota, Michigan, and Illinois (AOU 1998, Clapp et al. 1983). In the Pacifi c vagrants are known from California and the Hawaiian Islands (Hamilton et al. 2007, AOU 1998). Here we report vagrancy of Sandwich Terns to Isla Rasa, Gulf of California, Mexico, with our observations of single individuals in 1986 and again in 2008. -

Iucn Red Data List Information on Species Listed On, and Covered by Cms Appendices
UNEP/CMS/ScC-SC4/Doc.8/Rev.1/Annex 1 ANNEX 1 IUCN RED DATA LIST INFORMATION ON SPECIES LISTED ON, AND COVERED BY CMS APPENDICES Content General Information ................................................................................................................................................................................................................................ 2 Species in Appendix I ............................................................................................................................................................................................................................... 3 Mammalia ............................................................................................................................................................................................................................................ 4 Aves ...................................................................................................................................................................................................................................................... 7 Reptilia ............................................................................................................................................................................................................................................... 12 Pisces ................................................................................................................................................................................................................................................. -

Distribution and Abundance of Breeding Birds at Deception Island, South Shetland Islands, Antarctica, February to April 2000
Bó & Copello: Deception Island breeding birds’ distribution and abundance 39 DISTRIBUTION AND ABUNDANCE OF BREEDING BIRDS AT DECEPTION ISLAND, SOUTH SHETLAND ISLANDS, ANTARCTICA, FEBRUARY TO APRIL 2000 MARÍA SUSANA BÓ & SOFÍA COPELLO Universidad Nacional de Mar del Plata, Facultad de Ciencias Exactas y Naturales, Departamento de Biología, Laboratorio de Vertebrados, Funes 3350, 7600 Mar del Plata, Argentina ([email protected]) Received 20 September 2000, accepted 15 January 2001 SUMMARY BÓ, M.S. & COPELLO, S. 2000. Distribution and abundance of breeding birds at Deception Island, South Shetland Islands, Antarctica, February to April 2000. Marine Ornithology 29: 39–42. A survey of breeding birds during the brooding stage was carried out from February to April 2000 in the southern portion of Deception Island, South Shetland Islands, Antarctica. This island supports two Sites of Special Scien- tific Interest (SSSI Nos. 21 and 27). Nine species were found breeding in the study area: Chinstrap Penguin Pygoscelis antarctica (an estimated 6820 breeding pairs at two colonies surveyed), Pintado or Cape Petrel Daption capense (36), Wilson’s Storm Petrel Oceanites oceanicus (3), Antarctic Cormorant Phalacrocorax atriceps bransfieldensis (9), Greater Sheathbill Chionis alba (2), Subantarctic Skua Catharacta antarctica (4), South Polar Skua C. maccormicki (11), Kelp Gull Larus dominicanus (49) and Antarctic Tern Sterna vittata (5). Due to the increasing tourist activity at Deception Island, better information on the location and size of breeding populations is a particular requirement if effective precautionary conservation actions are to be taken. Key words: seabird censuses, Deception Island, Antarctica INTRODUCTION tal Protection to the Antarctic Treaty and the Convention on the Conservation of Antarctic Marine Living Resources (CCAMLR) Populations of most seabird species in Antarctica are stable or (Walton & Dingwall 1995). -

Nest Spacing in Elegant Terns: Hexagonal Packing Revisited Charles T
WESTERN BIRDS Volume 39, Number 2, 2008 NEST SPACING IN ELEGANT TERNS: HEXAGONAL PACKING REVISITED CHARLES T. COLLINS and MICHAEL D. TAYLOR, Department of Biological Sci- ences, California State University, Long Beach, California 90840 (current address of Taylor: Santiago Canyon College, 8045 East Chapman Ave., Orange, California 92869); [email protected] ABSTRACT: Within an important breeding colony in southern California, Elegant Terns (Thalasseus elegans) nest in one to several tightly packed clusters. Inter-nest distances within these clusters average 31.2 cm. This value is less than that reported for the larger-bodied Royal Tern (T. maximus) and Great Crested Tern (T. bergii). For Elegant Terns, the modal number of adjacent nests was six (range 5–7). This type of nest arrangement has been previously described as hexagonal packing and now appears to be typical of all Thalasseus terns for which data are available. Many seabirds nest in large, often traditional, colonies (Coulson 2002, Schreiber and Burger 2002). The ontogeny of annual colony formation has been reviewed by Kharitonov and Siegel-Causey (1988), and the evolution- ary processes which have led to coloniality have been considered by a number of authors (Lack 1968, Fischer and Lockley 1974, Wittenburger and Hunt 1985, Siegel-Causey and Kharitonov 1990, Coulson 2002). Seabird colonies may be rather loosely organized aggregations of breeding pairs of one to several species at a single site. At the other extreme, they may be dense, tightly packed, largely monospecific clusters where distances between nests are minimal. A graphic example of the latter is the dense clustering of nests recorded for several species of crested terns (Buckley and Buckley 1972, 2002, Hulsman 1977, Veen 1977, Symens and Evans 1993, Burness et al. -

Australian Painted Snipe in Bundaberg
BirdLife Bundaberg Volume 1 Number 1 March 2012 AUSTRALIAN PAINTED CLUB OUTINGS March 24-26th Boonaroo. Camping or single day outing. Meet Thabeban State School car park at 6am. SNIPE IN BUNDABERG Contact Leader Chris Barnes 0419723911 April 29th…………………..Goodnight Scrub NP. Meet North On the last club outing of 2011 we met in State Primary School 6am Leaders Eric Zillmann & Carl Moller the Bundaberg Botanic Gardens for a stroll 41551501 around. May 25-27th…………… Weekend campout to Middle While walking around the lake Potta spotted an Australian Creek. Details to be advised. Leader Bill Moorhead Painted Snipe in a shady drain. 41541177…… Soon we were all looking at four of these normally scarce birds. At the same time records were of Painted Snipe in several other locations around Australia. Contents The four birds in Bundaberg were all young females. The birds were sighted for around a fortnight. 1. Club outings Photo by Deane Lewis 2. Two campouts and a day in the Goodnight Scrub NP in the next three months . Paradise Riflebird .A rarity for a day 3. Lady Elliot Island 4. cont Lady Elliot Island BirdLife Bundaberg TWO CAMPOUTS AND A DAY IN A RARITY FOR A DAY. THE GOODNIGHT SCRUB NP IN Chris Barnes was doing an early morning THE NEXT THREE MONTHS. wader survey at the Bundaberg Port on Jan 6th found an interesting wader. Chris Boonaroo is the mecca of waders in our area. They phoned a few others and they were able to are there in their thousands and at this time of year see it as well. -

Breeding Biology of the Brown Noddy on Tern Island, Hawaii
Wilson Bull., 108(2), 1996, pp. 317-334 BREEDING BIOLOGY OF THE BROWN NODDY ON TERN ISLAND, HAWAII JENNIFER L. MEGYESI’ AND CURTICE R. GRIFFINS ABSTRACT.-we observed Brown Noddy (Anous stolidus pileatus) breeding phenology and population trends on Tern Island, French Frigate Shoals, Hawaii, from 1982 to 1992. Peaks of laying ranged from the first week in January to the first week in November; however, most laying occurred between March and September each year. Incubation length was 34.8 days (N = 19, SD = 0.6, range = 29-37 days). There were no differences in breeding pairs between the measurements of the first egg laid and successive eggs laid within a season. The proportion of light- and dark-colored chicks was 26% and 74%, respectively (N = 221) and differed from other Brown Noddy colonies studied in Atlantic and Pacific oceans. The length of time between clutches depended on whether the previous outcome was a failed clutch or a successfully fledged chick. Hatching, fledging, and reproductive success were significantly different between years. The subspecies (A. s. pihtus) differs in many aspects of its breeding biology from other colonies in the Atlantic and Pacific oceans, in regard to year-round occurrence at the colony, frequent renesting attempts, large egg size, proportion of light and dark colored chicks, and low reproductive success caused by in- clement weather and predation by Great Frigatebirds (Fregata minor). Received 31 Mar., 1995, accepted 5 Dec. 1995. The Brown Noddy (Anous stolidus) is the largest and most widely distributed of the tropical and subtropical tern species (Cramp 1985).