C. W. Cunningham. 2010
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Number 3, Spring 1998 Director’S Letter
Planning and planting for a better world Friends of the JC Raulston Arboretum Newsletter Number 3, Spring 1998 Director’s Letter Spring greetings from the JC Raulston Arboretum! This garden- ing season is in full swing, and the Arboretum is the place to be. Emergence is the word! Flowers and foliage are emerging every- where. We had a magnificent late winter and early spring. The Cornus mas ‘Spring Glow’ located in the paradise garden was exquisite this year. The bright yellow flowers are bright and persistent, and the Students from a Wake Tech Community College Photography Class find exfoliating bark and attractive habit plenty to photograph on a February day in the Arboretum. make it a winner. It’s no wonder that JC was so excited about this done soon. Make sure you check of themselves than is expected to seedling selection from the field out many of the special gardens in keep things moving forward. I, for nursery. We are looking to propa- the Arboretum. Our volunteer one, am thankful for each and every gate numerous plants this spring in curators are busy planting and one of them. hopes of getting it into the trade. preparing those gardens for The magnolias were looking another season. Many thanks to all Lastly, when you visit the garden I fantastic until we had three days in our volunteers who work so very would challenge you to find the a row of temperatures in the low hard in the garden. It shows! Euscaphis japonicus. We had a twenties. There was plenty of Another reminder — from April to beautiful seven-foot specimen tree damage to open flowers, but the October, on Sunday’s at 2:00 p.m. -

Leguminosae No Acervo Do Herbário Da Amazônia Meridional, Alta Floresta, Mato Grosso
LEGUMINOSAE NO ACERVO DO HERBÁRIO DA AMAZÔNIA MERIDIONAL, ALTA FLORESTA, MATO GROSSO José Martins Fernandes 1, Célia Regina Araújo Soares-Lopes 2,5 , Ricardo da Silva Ribeiro 3,5 & Dennis Rodrigues da Silva 4,5 1Professor do Curso de Licenciatura e Bacharelado em Ciências Biológicas, Universidade do Estado de Mato Grosso, Campus de Alta Floresta 2Professora Adjunta VI, da Faculdade de Ciências Biológicas e Agrárias de Alta Floresta, Universidade do Estado de Mato Grosso. ([email protected]) 3Graduando em Licenciatura e Bacharelado em Ciências Biológicas, Universidade do Estado de Mato Grosso, Campus de Alta Floresta 4Licenciado em Biologia, Universidade do Estado de Mato Grosso, Campus de Cáceres 5Herbário da Amazônia Meridional – HERBAM, Centro de Biodiversidade da Amazônia Meridional - CEBIAM Recebido em: 31/03/2015 – Aprovado em: 15/05/2015 – Publicado em: 01/06/2015 RESUMO Leguminosae é considerada a terceira maior família em número de espécies no mundo e a primeira no Brasil. Destaca-se nos diferentes domínios fitogeográficos brasileiros em riqueza e uso, como na Amazônia, porém, a Amazônia meridional ainda foi pouco amostrada. Desta forma, o presente trabalho teve como objetivo apresentar uma listagem para as espécies de Leguminosae depositadas no Herbário da Amazônia Meridional - HERBAM, Alta Floresta, Mato Grosso. As identificações dos espécimes de Leguminosae depositados na coleção do HERBAM foram conferidas com base em revisões taxonômicas e imagens do New York Botanical Garden em março de 2015. Leguminosae está representada por 153 espécies distribuídas em 59 gêneros. As espécies Apuleia leiocarpa (Vogel) J.F.Macbr. e Hymenaea parvifolia Huber estão ameaçadas de extinção, na categoria Vulnerável. -

A Rapid Biological Assessment of the Upper Palumeu River Watershed (Grensgebergte and Kasikasima) of Southeastern Suriname
Rapid Assessment Program A Rapid Biological Assessment of the Upper Palumeu River Watershed (Grensgebergte and Kasikasima) of Southeastern Suriname Editors: Leeanne E. Alonso and Trond H. Larsen 67 CONSERVATION INTERNATIONAL - SURINAME CONSERVATION INTERNATIONAL GLOBAL WILDLIFE CONSERVATION ANTON DE KOM UNIVERSITY OF SURINAME THE SURINAME FOREST SERVICE (LBB) NATURE CONSERVATION DIVISION (NB) FOUNDATION FOR FOREST MANAGEMENT AND PRODUCTION CONTROL (SBB) SURINAME CONSERVATION FOUNDATION THE HARBERS FAMILY FOUNDATION Rapid Assessment Program A Rapid Biological Assessment of the Upper Palumeu River Watershed RAP (Grensgebergte and Kasikasima) of Southeastern Suriname Bulletin of Biological Assessment 67 Editors: Leeanne E. Alonso and Trond H. Larsen CONSERVATION INTERNATIONAL - SURINAME CONSERVATION INTERNATIONAL GLOBAL WILDLIFE CONSERVATION ANTON DE KOM UNIVERSITY OF SURINAME THE SURINAME FOREST SERVICE (LBB) NATURE CONSERVATION DIVISION (NB) FOUNDATION FOR FOREST MANAGEMENT AND PRODUCTION CONTROL (SBB) SURINAME CONSERVATION FOUNDATION THE HARBERS FAMILY FOUNDATION The RAP Bulletin of Biological Assessment is published by: Conservation International 2011 Crystal Drive, Suite 500 Arlington, VA USA 22202 Tel : +1 703-341-2400 www.conservation.org Cover photos: The RAP team surveyed the Grensgebergte Mountains and Upper Palumeu Watershed, as well as the Middle Palumeu River and Kasikasima Mountains visible here. Freshwater resources originating here are vital for all of Suriname. (T. Larsen) Glass frogs (Hyalinobatrachium cf. taylori) lay their -

Endemic Wild Ornamental Plants from Northwestern Yunnan, China
HORTSCIENCE 40(6):1612–1619. 2005. have played an important role in world horti- culture and have been introduced to Western countries where they have been widely cul- Endemic Wild Ornamental Plants tivated. Some of the best known examples include Rhododendron, Primula, Gentiana, from Northwestern Yunnan, China Pedicularis, and Saussurea, which are all im- 1 portant genera in northwestern Yunnan (Chen Xiao-Xian Li and Zhe-Kun Zhou et al., 1989; Feng, 1983; Guan et al., 1998; Hu, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, P.R. 1990; Shi and Jin, 1999; Yang, 1956;). Many of China 650204 these ornamental species are endemic to small areas of northwestern Yunnan (e.g., Rhododen- Additional index words. horticultural potential dron russatum), therefore, their cultivation not Abstract. Northwestern Yunnan is situated in the southern part of the Hengduan Mountains, only provides for potential sources of income which is a complex and varied natural environment. Consequently, this region supports a generation, but also offers a potential form of great diversity of endemic plants. Using fi eld investigation in combination with analysis conservation management: these plants can of relevant literature and available data, this paper presents a regional ethnobotanical be used directly for their ornamental plant study of this area. Results indicated that northwestern Yunnan has an abundance of wild value or as genetic resources for plant breed- ornamental plants: this study identifi ed 262 endemic species (belonging to 64 genera and ing programs. The aims of current paper are 28 families) with potential ornamental value. The distinguishing features of these wild to describe the unique fl ora of northwestern plants, their characteristics and habitats are analyzed; the ornamental potential of most Yunnan and provide detailed information of plants stems from their wildfl owers, but some species also have ornamental fruits and those resources, in terms of their potential foliage. -

Phylogenetic Relationships in Rosaceae Inferred from Chloroplast Matk and Trnl-Trnf Nucleotide Sequence Data
Plant Syst. Evol. 231: 77±89 32002) Phylogenetic relationships in Rosaceae inferred from chloroplast matK and trnL-trnF nucleotide sequence data D. Potter1, F. Gao1, P. Esteban Bortiri1, S.-H. Oh1, and S. Baggett2 1Department of Pomology, University of California, Davis, USA 2Ph.D. Program Biology, Lehman College, City University of New York, New York, USA Received February 27, 2001 Accepted October 11, 2001 Abstract. Phylogenetic relationships in Rosaceae economically important fruits of temperate were studied using parsimony analysis of nucleo- regions is produced by members of this family, tide sequence data from two regions of the including species of Malus 3apples), Pyrus chloroplast genome, the matK gene and the trnL- 3pears), Prunus 3plums, peaches, cherries, trnF region. As in a previously published phylog- almonds, and apricots), Rubus 3raspberries), eny of Rosaceae based upon rbcL sequences, and Fragaria 3strawberries). The family also monophyletic groups were resolved that corre- includes many ornamentals, e.g., species of spond, with some modi®cations, to subfamilies Maloideae and Rosoideae, but Spiraeoideae were Rosa 3roses), Potentilla 3cinquefoil), and polyphyletic. Three main lineages appear to have Sorbus 3mountain ash). A variety of growth diverged early in the evolution of the family: 1) habits, fruit types, and chromosome numbers Rosoideae sensu stricto, including taxa with a base is found within the family 3Robertson 1974), chromosome number of 7 3occasionally 8); 2) which is traditionally divided into four sub- actinorhizal Rosaceae, a group of taxa that engage families on the basis of fruit type 3e.g., Schulze- in symbiotic nitrogen ®xation; and 3) the rest of the Menz 1964). -

Draft Plant Propagation Protocol
Plant Propagation Protocol for [Insert Species] ESRM 412 – Native Plant Production TAXONOMY Family Names Family Scientific Rosaceae Name: Family Common Rose Name: Scientific Names Genus: Physocarpus Species: malvaceus Species Authority: (Greene) Kuntz Variety: Sub-species: There are no recognized subspecies, varieties, or forms. Cultivar: Authority for N/A Variety/Sub- species: Common Neillia malvacea Greene Synonym(s) Neillia monogyna var. malvacea M. E. Jones (include full Neillia torreyi Hook. f. scientific names (e.g., Elymus Opulaster cordatus Rydb. glaucus Buckley), Opulaster pauciflorus Heller including variety or Opulaster pubescens Rydb. subspecies Physocarpus pauciflorus piper information) Spiraea opulifolia var. pauciflora T. & G. Spiraea pauciflora Nutt. Common Name(s): Species Code (as per PHMA5 USDA Plants database): GENERAL INFORMATION Geographical range (distribution maps for North America and Washington state) East of the Cascades, British Columbia to Oregon, east to Alberta and Montana, and south to Utah and Wyoming Ecological Soil textures found on ninebark sites range from sandy loams to distribution silty clay loams. Ash layers have been found on some soils in (ecosystems it central Idaho. Surface soil acidity range from pH 5.5 to 7.1. occurs in, etc): The average duff layer is 2.6 inches (6.5 cm) with an effective rooting depth generally at 15 inches (38 cm). (7) Climate and It has been found on moist slopes and streamsides 5,250 to 10,000 feet (1,600- elevation range 3,000 m) Local habitat and Ninebark is most commonly found on hillsides, canyons, and grasslands on abundance; may ponderosa pine and Douglas fir site, in mountain-brush, aspen, and mixed-conifer include commonly woodlands. -

Djvu Document
BULL. BOT. SURV. INDIA VoL 21, Nos. 1-4 : pp. 35-67, 1979 COMMEMORATIVE GENERIC NAMES OF FLOWERING PLANTS IN INDIA Botanical Surq of India, Howrah ABSTRACT Commemorative generic names of plants are given in honour of well known botanists and patrons of science for the contributions they made for the cause of botany. The names are given in alphabetical order with their families and one example under each gcnw. A brief biographical sketch of the person is also given. Plants named after botanists and eminent persons form one continuous historical sequence in the development of Natural history. INTRODUCTION In 1867 Alphonse de Candolle, son of A. P. Naming of plants and animals using two- De Candolle with his legal background first word names is as old as human civilization proposed the rules of nomenclature in "Lois and is commonly seen in all languages. In de la Nomenclature Botanique" (1867). In I 753 Carl ~innaeusin his Species Plantarum the Vienna botanical congress (Igo j), these consistently used the two-word system in rules were officially designated as "Interna- Latin for about 5900 Linnean specific epi- tional Rules of Botanical Nomenclature" and thets which was hailed as the binomial since 1952 it is called as "International Code system of nomenclature. In the 18th of Botanical Nomenclature" Plant names century in Europe, Latin was the accepted named after botanists and eminent persous language of scholarship and common form one continuous historical sequence in medium of communication cutting across the development of Natural history. Gen- national boundaries. According to the eric names are given in alphabetical order rules of naming mostly botanical generic and under the first genus Abelia R. -
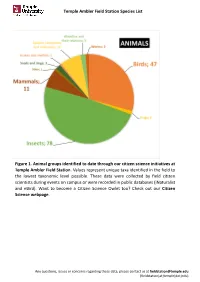
Temple Ambler Field Station Species List Figure 1. Animal Groups Identified to Date Through Our Citizen Science Initiatives at T
Temple Ambler Field Station Species List Figure 1. Animal groups identified to date through our citizen science initiatives at Temple Ambler Field Station. Values represent unique taxa identified in the field to the lowest taxonomic level possible. These data were collected by field citizen scientists during events on campus or were recorded in public databases (iNaturalist and eBird). Want to become a Citizen Science Owlet too? Check out our Citizen Science webpage. Any questions, issues or concerns regarding these data, please contact us at [email protected] (fieldstation[at}temple[dot]edu) Temple Ambler Field Station Species List Figure 2. Plant diversity identified to date in the natural environments and designed gardens of the Temple Ambler Field Station and Ambler Arboretum. These values represent unique taxa identified to the lowest taxonomic level possible. Highlighted are 14 of the 116 flowering plant families present that include 524 taxonomic groups. A full list can be found in our species database. Cultivated specimens in our Greenhouse were not included here. Any questions, issues or concerns regarding these data, please contact us at [email protected] (fieldstation[at}temple[dot]edu) Temple Ambler Field Station Species List database_title Temple Ambler Field Station Species List last_update 22October2020 description This database includes all species identified to their lowest taxonomic level possible in the natural environments and designed gardens on the Temple Ambler campus. These are occurrence records and each taxon is only entered once. This is an occurrence record, not an abundance record. IDs were performed by senior scientists and specialists, as well as citizen scientists visiting campus. -

Fruit Trees and Useful Plants in Amazonian Life (2011)
FAO TECHNICAL PAPERS NON-WOOD FOREST PRODUCTS 1. Flavours and fragrances of plant origin (1995) 2. Gum naval stores: turpentine and rosin from pine resin (1995) 3. Report of the International Expert Consultation on Non-Wood Forest Products (1995) 4. Natural colourants and dyestuffs (1995) 5. Edible nuts (1995) 6. Gums, resins and latexes of plant origin (1995) 7. Non-wood forest products for rural income and sustainable forestry (1995) 8. Trade restrictions affecting international trade in non-wood forest products (1995) 9. Domestication and commercialization of non-timber forest products in agroforestry systems (1996) 10. Tropical palms (1998) 11. Medicinal plants for forest conservation and health care (1997) 12. Non-wood forest products from conifers (1998) 13. Resource assessment of non-wood forest products Experience and biometric principles (2001) 14. Rattan – Current research issues and prospects for conservation and sustainable development (2002) 15. Non-wood forest products from temperate broad-leaved trees (2002) 16. Rattan glossary and Compendium glossary with emphasis on Africa (2004) 17. Wild edible fungi – A global overview of their use and importance to people (2004) 18. World bamboo resources – A thematic study prepared in the framework of the Global Forest Resources Assessment 2005 (2007) 19. Bees and their role in forest livelihoods – A guide to the services provided by bees and the sustainable harvesting, processing and marketing of their products (2009) 20. Fruit trees and useful plants in Amazonian life (2011) The -

Neillia Includes Stephanandra (Rosaceae)
Neillia Includes Stephanandra (Rosaceae) Sang-Hun Oh Department of Biology, 139 Biological Sciences, Box 90338, Duke University, Durham, North Carolina 27708, U.S.A. [email protected] ABSTRACT . Recent phylogenetic analyses of tribe Furthermore, Physocarpus is quite morphologically Neillieae in Rosaceae, which comprises Neillia, distinct from Neillia and Stephanandra in having Physocarpus, and Stephanandra, based on both stellate trichomes, corymbose inflorescences, and nuclear and chloroplast DNA sequences revealed that inflated follicular fruits dehiscent along both ventral Neillia and Stephanandra together form a strongly and dorsal sutures. supported monophyletic group and that species of Neillia and Stephanandra have been distinguished Stephanandra are nested within Neillia. The close based on characters in fruits and seeds (Vidal, 1963; relationship between Neillia and Stephanandra is also Cullen, 1971), but my examination of herbarium supported by leaf morphology, inflorescence type, and specimens, including the type collections, indicates carpel number. In order to better reflect the that the two genera are not clearly separable with evolutionary relationships among species of Neillia these characters. Vidal (1963), in his revision of and Stephanandra, based on molecular phylogenetic Neillia, stated that follicles of Stephanandra in- and morphological evidence, Stephanandra is united completely dehisce at maturity, whereas those of with Neillia, and three traditionally recognized Neillia are completely dehiscent. However, the mature species and a hybrid of Stephanandra are transferred follicles of some species of Neillia, such as N. sinensis to Neillia. This treatment results in new combinations Oliver, N. thibetica Bureau & Franchet, and N. uekii as follows: N. hanceana (Kuntze) S. Oh, N. incisa Nakai, are not completely dehiscent, with the result (Thunberg) S. -

Anatomía De Madera En 31 Especies De La Subfamilia Mimosoideae (Leguminosae) En Venezuela
ANATOMÍA DE MADERA EN 31 ESPECIES DE LA SUBFAMILIA MIMOSOIDEAE (LEGUMINOSAE) EN VENEZUELA Palabras clave: Mimosoideae, anatomía de la madera, xilema secundario, identificación. Williams J. León H.1 Key Words: Mimosoideae, wood anatomy, secondary xylem, identification. RESUMEN se logró elaborar una clave para la identificación de las especies estudiadas. El presente trabajo tiene como objetivo estudiar la anatomía de la madera de 31 especies, correspon- ABSTRACT dientes a 17 géneros, de la subfamilia Mimosoideae (Leguminosae), procedentes de diferentes regiones This paper is about the wood anatomy of 31 species, geográficas de Venezuela. Se estudiaron entre uno belonging to 17 genera, of the Mimosoideae y cinco individuos por especie. Las descripciones subfamily (Leguminosae), proceeding from different se realizaron siguiendo lo estipulado por el IAWA geographical regions of Venezuela. For each species, Committee (1989). Las especies estudiadas se pue- one to five individuals were studied. The descriptions den dividir en dos grandes grupos, de acuerdo a la were realized according to the IAWA Committee (1989). presencia o ausencia de fibras septadas. El género The studied species may be divided in two groups Inga se caracterizó porque todas las especies es- according to the presence or absence of septate fibers. tudiadas presentaron fibras septadas, mientras que All species of Inga showed septate fibers, whereas Albizia y Enterolobium presentaron especies con fi- Albizia and Enterolobium included species with bras septadas y especies con fibras no septadas. Las septate fibers and also species with non-septate fibers. características cuantitativas de los vasos y el ancho The quantitative characteristics of the vessels and the de radio mostraron suficiente variación como para width of rays showed sufficient variation as to be ser consideradas características importantes desde considered important characteristics from a taxonomic el punto de vista taxonómico. -

05 Kloster Et Al Pg391a402 PB.Indd
Rev. bras. paleontol. 18(3):391-402, Setembro/Dezembro 2015 © 2015 by the Sociedade Brasileira de Paleontologia doi: 10.4072/rbp.2015.3.05 NEW RECORD OF FABACEAE FOSSIL WOODS FROM THE SOLIMÕES FORMATION (MIOCENE), ACRE BASIN, AMAZON, BRAZIL ADRIANA KLOSTER Centro de Ecología Aplicada del Litoral, Área de Paleontología, CECOAL-CONICET, Ruta 5, km 2,5, CP. 3400, Corrientes, Argentina. [email protected] SILVIA GNAEDINGER Centro de Ecología Aplicada del Litoral, Área de Paleontología, CECOAL-CONICET, Ruta 5, km 2,5, CP. 3400. FACENA UNNE, Casilla correo 128, CP. 3400, Corrientes, Argentina. [email protected] KAREN ADAMI-RODRIGUES & CAMILE URBAN Núcleo de Estudos em Paleontologia e Estratigrafi a (NEPALE), Centro das Engenharias, UFPel, Praça Domingos Rodrigues, 2, 96010-440, Pelotas, RS. Brazil. [email protected], [email protected] ABSTRACT – Detailed descriptions are provided for specimens of a new genus and species of Mimosoideae and Swartzioxylon naraense Delteil-Desneux emended (Papilionoideae) from the Solimões Formation, Miocene, Acre Basin, Brazil. Anatomical characteristics of the woods show a resemblance with the extant genera Zygia P. Browne and Swartzia Schreb, respectively. These genera currently have a wide distribution in Brazil and throughout the Amazon region; their presence in the outcrops of the Solimões Formation supports the idea that these genera might have been important components and indicates their importance in the fossil record, both to establish its distribution and evolution from the past to present of Amazon fl ora. Key words: wood anatomy, Mimosoideae, Papilionoideae, Neogene. RESUMO – Descrições detalhadas são fornecidas para os espécimes de um novo gênero e espécie de Mimosoideae e Swartzioxylon naraense Delteil-Desneux emend.