The Fossil Record of Camelids Demonstrates a Late Divergence Between Bactrian Camel and Dromedary
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Wild Or Bactrian Camel French: German: Wildkamel Spanish: Russian: Dikiy Verblud Chinese
1 of 4 Proposal I / 7 PROPOSAL FOR INCLUSION OF SPECIES ON THE APPENDICES OF THE CONVENTION ON THE CONSERVATION OF MIGRATORY SPECIES OF WILD ANIMALS A. PROPOSAL: Inclusion of the Wild camel Camelus bactrianus in Appendix I of the Convention on the Conservation of Migratory Species of Wild Animals: B. PROPONENT: Mongolia C. SUPPORTING STATEMENT 1. Taxon 1.1. Classis: Mammalia 1.2. Ordo: Tylopoda 1.3. Familia: Camelidae 1.4. Genus: Camelus 1.5. Species: Camelus bactrianus Linnaeus, 1758 1.6. Common names: English: Wild or Bactrian camel French: German: Wildkamel Spanish: Russian: Dikiy verblud Chinese: 2. Biological data 2.1. Distribution Wild populations are restricted to 3 small, remnant populations in China and Mongolia:in the Taklamakan Desert, the deserts around Lop Nur, and the area in and around region A of Mongolia’s Great Gobi Strict Protected Area (Reading et al 2000). In addition, there is a small semi-captive herd of wild camels being maintained and bred outside of the Park. 2.2. Population Surveys over the past several decades have suggested a marked decline in wild bactrian camel numbers and reproductive success rates (Zhirnov and Ilyinsky 1986, Anonymous 1988, Tolgat and Schaller 1992, Tolgat 1995). Researchers suggest that fewer than 500 camels remain in Mongolia and that their population appears to be declining (Xiaoming and Schaller 1996). Globally, scientists have recently suggested that less than 900 individuals survive in small portions of Mongolia and China (Tolgat and Schaller 1992, Hare 1997, Tolgat 1995, Xiaoming and Schaller 1996). However, most of the population estimates from both China and Mongolia were made using methods which preclude rigorous population estimation. -
![About Pigs [PDF]](https://docslib.b-cdn.net/cover/0911/about-pigs-pdf-50911.webp)
About Pigs [PDF]
May 2015 About Pigs Pigs are highly intelligent, social animals, displaying elaborate maternal, communicative, and affiliative behavior. Wild and feral pigs inhabit wide tracts of the southern and mid-western United States, where they thrive in a variety of habitats. They form matriarchal social groups, sleep in communal nests, and maintain close family bonds into adulthood. Science has helped shed light on the depths of the remarkable cognitive abilities of pigs, and fosters a greater appreciation for these often maligned and misunderstood animals. Background Pigs—also called swine or hogs—belong to the Suidae family1 and along with cattle, sheep, goats, camels, deer, giraffes, and hippopotamuses, are part of the order Artiodactyla, or even-toed ungulates.2 Domesticated pigs are descendants of the wild boar (Sus scrofa),3,4 which originally ranged through North Africa, Asia and Europe.5 Pigs were first domesticated approximately 9,000 years ago.6 The wild boar became extinct in Britain in the 17th century as a result of hunting and habitat destruction, but they have since been reintroduced.7,8 Feral pigs (domesticated animals who have returned to a wild state) are now found worldwide in temperate and tropical regions such as Australia, New Zealand, and Indonesia and on island nations, 9 such as Hawaii.10 True wild pigs are not native to the New World.11 When Christopher Columbus landed in Cuba in 1493, he brought the first domestic pigs—pigs who subsequently spread throughout the Spanish West Indies (Caribbean).12 In 1539, Spanish explorers brought pigs to the mainland when they settled in Florida. -

Bactrian Camel, Two-Humped Camel
Camelus ferus/bactrianus Common name: Bactrian camel, two-humped camel Local name: Havtagai (Mongolian), Wildkamel (German), Jya nishpa yapung (Ladakhi) Classification: Kingdom: Animalia Phylum: Chordata Class: Mammalia Order: Artiodactyla Family: Camelidae Genus: Camelus Species: ferus/bactrianus Profile: The scientific name of the wild Bactrian camel is Camelus ferus, while the domesticated form is called Camelus bactrianus. The distinctive feature of the animal is that it is two-humped whereas the Dromedary camel has a single hump. DNA tests have revealed that there are two or three distinct genetic differences and about 3% base difference between the wild and domestic populations of Bactrian camels. They also differ physically. The wild Bactrian camel is smaller and slender than the domestic breed. The wild camels have a sandy gray- brown coat while the domestic ones have a dark brown coat. The predominant difference between them however is the shape of the humps. While that of the wild camel are small and pyramid-like, those of the domestic ones are large and irregular. The face of a Bactrian camel is long and triangular with a split upper lip. The Bactrian camel is highly adapted to surviving the cold desert climate. Each foot has an undivided sole with two large toes that can spread wide apart for walking on sand. The ears and nose are lined with hair to protect against sand and the muscular nostrils can be closed during sandstorms. The eyes are protected from sand and debris by a double layer of long eyelashes while bushy eyebrows give protection from the sun. It grows a thick shaggy coat during winter, which is shed very rapidly in spring to give the animal a shorn look. -

Bison Literature Review Biology
Bison Literature Review Ben Baldwin and Kody Menghini The purpose of this document is to compare the biology, ecology and basic behavior of cattle and bison for a management context. The literature related to bison is extensive and broad in scope covering the full continuum of domestication. The information incorporated in this review is focused on bison in more or less “wild” or free-ranging situations i.e.., not bison in close confinement or commercial production. While the scientific literature provides a solid basis for much of the basic biology and ecology, there is a wealth of information related to management implications and guidelines that is not captured. Much of the current information related to bison management, behavior (especially social organization) and practical knowledge is available through local experts, current research that has yet to be published, or popular literature. These sources, while harder to find and usually more localized in scope, provide crucial information pertaining to bison management. Biology Diet Composition Bison evolutional history provides the basis for many of the differences between bison and cattle. Bison due to their evolution in North America ecosystems are better adapted than introduced cattle, especially in grass dominated systems such as prairies. Many of these areas historically had relatively low quality forage. Bison are capable of more efficient digestion of low-quality forage then cattle (Peden et al. 1973; Plumb and Dodd 1993). Peden et al. (1973) also found that bison could consume greater quantities of low protein and poor quality forage then cattle. Bison and cattle have significant dietary overlap, but there are slight differences as well. -

Last Interglacial (MIS 5) Ungulate Assemblage from the Central Iberian Peninsula: the Camino Cave (Pinilla Del Valle, Madrid, Spain)
Palaeogeography, Palaeoclimatology, Palaeoecology 374 (2013) 327–337 Contents lists available at SciVerse ScienceDirect Palaeogeography, Palaeoclimatology, Palaeoecology journal homepage: www.elsevier.com/locate/palaeo Last Interglacial (MIS 5) ungulate assemblage from the Central Iberian Peninsula: The Camino Cave (Pinilla del Valle, Madrid, Spain) Diego J. Álvarez-Lao a,⁎, Juan L. Arsuaga b,c, Enrique Baquedano d, Alfredo Pérez-González e a Área de Paleontología, Departamento de Geología, Universidad de Oviedo, C/Jesús Arias de Velasco, s/n, 33005 Oviedo, Spain b Centro Mixto UCM-ISCIII de Evolución y Comportamiento Humanos, C/Sinesio Delgado, 4, 28029 Madrid, Spain c Departamento de Paleontología, Facultad de Ciencias Geológicas, Universidad Complutense de Madrid, Ciudad Universitaria, 28040 Madrid, Spain d Museo Arqueológico Regional de la Comunidad de Madrid, Plaza de las Bernardas, s/n, 28801-Alcalá de Henares, Madrid, Spain e Centro Nacional de Investigación sobre la Evolución Humana (CENIEH), Paseo Sierra de Atapuerca, s/n, 09002 Burgos, Spain article info abstract Article history: The fossil assemblage from the Camino Cave, corresponding to the late MIS 5, constitutes a key record to un- Received 2 November 2012 derstand the faunal composition of Central Iberia during the last Interglacial. Moreover, the largest Iberian Received in revised form 21 January 2013 fallow deer fossil population was recovered here. Other ungulate species present at this assemblage include Accepted 31 January 2013 red deer, roe deer, aurochs, chamois, wild boar, horse and steppe rhinoceros; carnivores and Neanderthals Available online 13 February 2013 are also present. The origin of the accumulation has been interpreted as a hyena den. Abundant fallow deer skeletal elements allowed to statistically compare the Camino Cave fossils with other Keywords: Early Late Pleistocene Pleistocene and Holocene European populations. -

Chapter 1 - Introduction
EURASIAN MIDDLE AND LATE MIOCENE HOMINOID PALEOBIOGEOGRAPHY AND THE GEOGRAPHIC ORIGINS OF THE HOMININAE by Mariam C. Nargolwalla A thesis submitted in conformity with the requirements for the degree of Doctor of Philosophy Graduate Department of Anthropology University of Toronto © Copyright by M. Nargolwalla (2009) Eurasian Middle and Late Miocene Hominoid Paleobiogeography and the Geographic Origins of the Homininae Mariam C. Nargolwalla Doctor of Philosophy Department of Anthropology University of Toronto 2009 Abstract The origin and diversification of great apes and humans is among the most researched and debated series of events in the evolutionary history of the Primates. A fundamental part of understanding these events involves reconstructing paleoenvironmental and paleogeographic patterns in the Eurasian Miocene; a time period and geographic expanse rich in evidence of lineage origins and dispersals of numerous mammalian lineages, including apes. Traditionally, the geographic origin of the African ape and human lineage is considered to have occurred in Africa, however, an alternative hypothesis favouring a Eurasian origin has been proposed. This hypothesis suggests that that after an initial dispersal from Africa to Eurasia at ~17Ma and subsequent radiation from Spain to China, fossil apes disperse back to Africa at least once and found the African ape and human lineage in the late Miocene. The purpose of this study is to test the Eurasian origin hypothesis through the analysis of spatial and temporal patterns of distribution, in situ evolution, interprovincial and intercontinental dispersals of Eurasian terrestrial mammals in response to environmental factors. Using the NOW and Paleobiology databases, together with data collected through survey and excavation of middle and late Miocene vertebrate localities in Hungary and Romania, taphonomic bias and sampling completeness of Eurasian faunas are assessed. -

An Educator's Resource to Texas Mammal Skulls and Skins
E4H-014 11/17 An Educator’s Resource to Texas Mammal Skulls and Skins for use in 4-H Wildlife Programs and FFA Wildlife Career Development Events By, Denise Harmel-Garza Program Coordinator I, Texas A&M AgriLife Extension Service, 4-H Photographer and coauthor, Audrey Sepulveda M.Ed. Agricultural Leadership, Education and Communications, Texas A&M University College Station, Texas 2017 “A special thanks to the Biodiversity Research and Teaching Collections at Texas A&M University for providing access to their specimens.” Texas A&M AgriLife Extension provides equal opportunities in its programs and employment to all persons, regardless of race, color, sex, religion, national origin, disability, age, genetic information, veteran status, sexual orientation, or gender identity. The Texas A&M University System, U.S. Department of Agriculture, and the County Commissioners Courts of Texas Cooperating. Introduction Texas youth that participate in wildlife programs may be asked to identify a skull, skin, scat, tracks, etc. of an animal. Usually, educators must find this information and assemble pictures of skulls and skins from various sources. They also must ensure that what they find is relevant and accurate. Buying skulls and skins to represent all Texas mammals is costly. Most educators cannot afford them, and if they can, maintaining these collections over time is problematic. This study resource will reduce the time teachers across the state need to spend searching for information and allow them more time for presenting the material to their students. This identification guide gives teachers and students easy access to information that is accurate and valuable for learning to identify Texas mammals. -

1 BOARD of ANIMAL HEALTH Subpart 2 Chapter 12 Sheep And
BOARD OF ANIMAL HEALTH Subpart 2 Chapter 12 Sheep and Goats 109 All sheep and goats, except those for immediate slaughter shall be accompanied by an official certificate of veterinary inspection (OCVI) and shall comply with the following: 1. Intact sheep and goats require individual identification by an official USDA Scrapie eartag, brand, or tattoo recorded on the OCVI. 2. “I certify these animals are free of clinical signs of the diseases contagious footrot, keratoconjunctivitis, contagious ecthyma (Orf), scabies and lice and that the sexually intact animals represented on this form are not known to be scrapie- positive, suspect, high risk, or exposed, and did not originate from a known infected, source, exposed, or noncompliant flock.” 3. When originating from an area known to have scabies, must be dipped within ten (10) days immediately preceding the date of entry in an USDA approved dip, and maintained on absolutely clean premises until delivered to the final destination. Dairy goats and dairy sheep maintained separate from other sheep and goats are exempt from dipping when certified free of scabies on OCVI. 4. Dairy goats and dairy sheep over 6 months of age must be negative to an official tuberculin test and an official brucellosis test made within 30 days immediately preceding date of entry. 5. All sheep and goats for immediate slaughter shall be consigned to a recognized slaughtering establishment on either an OCVI or permit or waybill or inspection certification from federally inspected stockyards. In either instance, a copy shall accompany sheep and goats and a copy shall be forwarded to the State Veterinarian of Mississippi. -

6. Petronio Di Stefano Pandolfi Salari.Indd
Bollettino del Museo Civico di Storia Naturale di Verona, 38, 2014 Geologia Paleontologia Preistoria: 103-116 The Late Pleistocene mammal fauna from Montemerano - Manciano (Grosseto, central Italy) CARMELO PETRONIO*, GIUSEPPE DI STEFANO*, LUCA PANDOLFI**, LEONARDO SALARI* *Dipartimento di Scienze della Terra, “Sapienza” Università di Roma **Dipartimento di Scienze Geologiche, Università degli Studi Roma Tre Abstract The Late Pleistocene mammal fauna from Montemerano – Manciano (Grosseto, central Italy) – Several fossil mammal remains have been recovered in a karst cavity near Montemerano (Manciano, Grosseto, Central Italy). The cavity was developed in the upper travertine deposits outcropping in the area and chronologically referred to a time span between the late Middle and the Late Pleistocene. The paleontological analysis of the mammal remains allows to recognize different taxa as Panthera spelaea, Canis lupus, Stephanorhi- nus cf. S. hemitoechus, Equus ferus, Sus scrofa, Bison sp., Bos primigenius, Megaloceros giganteus, Cervus elaphus, Dama dama and some coprolites of Hyaenidae. However, only the occurrence of an evolved form of Dama dama in the assemblage provides useful biochronological information. The fallow deer is represented by several remains, in particular by a fragmentary mandible with the molars. The mandible shows morphological and morphometrical features closer to the Late Pleistocene populations of the fallow deer than to the archaic Dama clactoniana and D. dama tiberina. The evolved forms of the fallow deer were typical and abundant during the early Late Pleistocene. Nevertheless, they were very rare during the late Late Pleistocene and became extinct in Italy during the Last Glacial Maximum, probably due to the climate changes. Finally, a preliminary analysis of the sediment including the mammal remains shows a significant decrease of the volcanic elements coming from the Vulsino Volcanic District (VVD). -

Pleistocene Mammals from Extinction Cave, Belize
Canadian Journal of Earth Sciences Pleistocene Mammals From Extinction Cave, Belize Journal: Canadian Journal of Earth Sciences Manuscript ID cjes-2018-0178.R3 Manuscript Type: Article Date Submitted by the 04-May-2019 Author: Complete List of Authors: Churcher, C.S.; University of Toronto, Zoology Central America, Pleistocene, Fauna, Vertebrate Palaeontology, Keyword: Limestone cave Is the invited manuscript for consideration in a Special Not applicableDraft (regular submission) Issue? : https://mc06.manuscriptcentral.com/cjes-pubs Page 1 of 43 Canadian Journal of Earth Sciences 1 1 PLEISTOCENE MAMMALS FROM EXTINCTION CAVE, BELIZE 2 by C.S. CHURCHER1 Draft 1Department of Zoology, University of Toronto, Toronto, Ontario Canada M5S 2C6 and 322-240 Dallas Rd., Victoria, British Columbia, Canada V8V 4X9 (corresponding address): e-mail [email protected] https://mc06.manuscriptcentral.com/cjes-pubs Canadian Journal of Earth Sciences Page 2 of 43 2 4 5 ABSTRACT. A small mammalian fauna is recorded from Extinction Cave (also called Sibun 6 Cave), east of Belmopan, on the Sibun River, Belize, Central America. The animals recognized 7 are armadillo (†Dasypus bellus), American lion (†Panthera atrox), jaguar (P. onca), puma or 8 mountain lion (Puma concolor), Florida spectacled bear (†Tremarctos floridanus), javelina or 9 collared peccary (Pecari tajacu), llama (Camelidae indet., ?†Palaeolama mirifica), red brocket 10 deer (Mazama americana), bison (Bison sp.) and Mexican half-ass (†Equus conversidens), and 11 sabre-tooth cat († Smilodon fatalis) may also be represented (‘†’ indicates an extinct taxon). 12 Bear and bison are absent from the region today. The bison record is one of the more southernly 13 known. The bear record is almost the mostDraft westerly known and a first for Central America. -
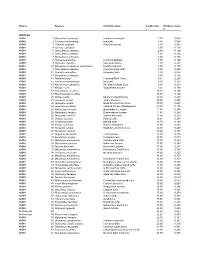
PDF File Containing Table of Lengths and Thicknesses of Turtle Shells And
Source Species Common name length (cm) thickness (cm) L t TURTLES AMNH 1 Sternotherus odoratus common musk turtle 2.30 0.089 AMNH 2 Clemmys muhlenbergi bug turtle 3.80 0.069 AMNH 3 Chersina angulata Angulate tortoise 3.90 0.050 AMNH 4 Testudo carbonera 6.97 0.130 AMNH 5 Sternotherus oderatus 6.99 0.160 AMNH 6 Sternotherus oderatus 7.00 0.165 AMNH 7 Sternotherus oderatus 7.00 0.165 AMNH 8 Homopus areolatus Common padloper 7.95 0.100 AMNH 9 Homopus signatus Speckled tortoise 7.98 0.231 AMNH 10 Kinosternon subrabum steinochneri Florida mud turtle 8.90 0.178 AMNH 11 Sternotherus oderatus Common musk turtle 8.98 0.290 AMNH 12 Chelydra serpentina Snapping turtle 8.98 0.076 AMNH 13 Sternotherus oderatus 9.00 0.168 AMNH 14 Hardella thurgi Crowned River Turtle 9.04 0.263 AMNH 15 Clemmys muhlenbergii Bog turtle 9.09 0.231 AMNH 16 Kinosternon subrubrum The Eastern Mud Turtle 9.10 0.253 AMNH 17 Kinixys crosa hinged-back tortoise 9.34 0.160 AMNH 18 Peamobates oculifers 10.17 0.140 AMNH 19 Peammobates oculifera 10.27 0.140 AMNH 20 Kinixys spekii Speke's hinged tortoise 10.30 0.201 AMNH 21 Terrapene ornata ornate box turtle 10.30 0.406 AMNH 22 Terrapene ornata North American box turtle 10.76 0.257 AMNH 23 Geochelone radiata radiated tortoise (Madagascar) 10.80 0.155 AMNH 24 Malaclemys terrapin diamondback terrapin 11.40 0.295 AMNH 25 Malaclemys terrapin Diamondback terrapin 11.58 0.264 AMNH 26 Terrapene carolina eastern box turtle 11.80 0.259 AMNH 27 Chrysemys picta Painted turtle 12.21 0.267 AMNH 28 Chrysemys picta painted turtle 12.70 0.168 AMNH 29 -

Preliminary Post-Cranial Metric Analysis of Mammoths from the Hot Springs Mammoth Site, South Dakota
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Transactions of the Nebraska Academy of Sciences and Affiliated Societies Nebraska Academy of Sciences 1977 Preliminary Post-Cranial Metric Analysis Of Mammoths From The Hot Springs Mammoth Site, South Dakota Barbara Lee Dutrow Chadron State College Follow this and additional works at: https://digitalcommons.unl.edu/tnas Dutrow, Barbara Lee, "Preliminary Post-Cranial Metric Analysis Of Mammoths From The Hot Springs Mammoth Site, South Dakota" (1977). Transactions of the Nebraska Academy of Sciences and Affiliated Societies. 445. https://digitalcommons.unl.edu/tnas/445 This Article is brought to you for free and open access by the Nebraska Academy of Sciences at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Transactions of the Nebraska Academy of Sciences and Affiliated Societiesy b an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. COLLEGIATE SECTION PRELIMINARY POST-CRANIAL METRIC ANALYSIS OF MAMMOTHS FROM THE HOT SPRINGS MAMMOTH SITE, SOUTH DAKOTA BARBARA LEE DUTROW Department of Earth Sciences Chadron State College, Chadron, Nebraska 69337 Salvage investigations of a Karst depression containing more than but is not currently available. nine Late Pleistocene mammoth were conducted during approxi mately twenty field days in the summers of 1974 and 1975. The deposit contains a local death assemblage of a mammoth population. METHODOLOGY A post-cranial metric analysis has been conducted on the fossil elephant remains. Excavation of the site followed standard archaeological and paleontological techniques as applied at the following t t t sites: Murray Springs, Lehner Ranch, Boney Springs, and Hudson-Meng. Bones were mapped in situ, both vertically INTRODUCTION and horizontally, as encountered in the fill.