Initial Identification of UDP-Glucose Dehydrogenase As a Prognostic
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Critical Evaluation of Gene Expression Changes in Human Tissues In
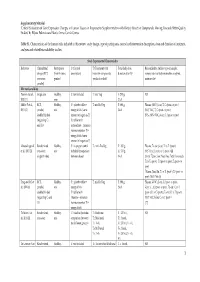
Supplementary Material ‘Critical Evaluation of Gene Expression Changes in Human Tissues in Response to Supplementation with Dietary Bioactive Compounds: Moving Towards Better-Quality Studies’ by Biljana Pokimica and María-Teresa García-Conesa Table S1. Characteristics of the human trials included in this review: study design, type of participants, control and intervention description, dose and duration of treatment, analyses and related bioavailability studies. Study Experimental Characteristics Reference Clinical trial Participants C (Control T (Treatment with Total daily dose, Bioavailability studies: type of sample, design (RCT, (health status, description) bioactive compounds, duration (d or h)1 compounds and (or) metabolites analysed, crossover, gender) products or diet) main results2 parallel) Mix meals and diets Persson I et al., Single arm Healthy, C: not included T: mix Veg T: 250 g, NR 2000 [1] men 21 d Møller P et al., RCT, Healthy, C1: placebo tablet + T: mix FruVeg T: 600 g, Plasma: (NS↑) β-car, T, C2 (post- vs pre-) 2003 [2] parallel, mix energy drink (same 24 d (NC) VitC, T, C2 (post- vs pre-) double blinded amount of sugars as T) (NS↓, 69%) VitC, β-car, C1 (post- vs pre-) (regarding C1 C2: tablet with and C2) antioxidants + minerals (same amount as T) + energy drink (same amount of sugars as T) Almendingen K Randomized, Healthy, C: no proper control T1,2: mix FruVeg T1: 300 g, Plasma: ↑α-car, β-car, T2 vs T1 (post-) et al., 2005 [3] crossover, mix included (comparison T2: 750 g, (NS↑) Lyc, Lut, T2 vs T1 (post-) [4] single -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Download Product Insert (PDF)
PRODUCT INFORMATION SETD3 (human, recombinant) Item No. 27355 Overview and Properties Synonyms: Actin Histidine Methyltransferase, Actin Histidine N-Methyltransferase, C14orf154, Chromosome 14 Open Reading Frame 154, HSETD3, SET Doman-Containing 3, SET Domain-containing Protein 3 Source: Active recombinant N-terminal His-tagged SETD3 expressed in E. coli Amino Acids: 2-594 (full length) Uniprot No.: Q86TU7 Molecular Weight: 69.2 kDa Storage: -80°C (as supplied) Stability: ≥1 year Purity: batch specific (≥80% estimated by SDS-PAGE) Supplied in: 50 mM HEPES, pH 8.0, with 150 mM sodium chloride and 10% glycerol Protein Concentration: batch specific mg/ml Activity: batch specific U/ml Specific Activity: batch specific U/mg Unit Definition: nmol/min/mg. One unit is defined as the amount of enzyme required to transfer one methyl group to actin peptide per minute using 8 µM actin peptide (LKYPIEHGIVTNWDDMEKIW-amide) at 37°C in Cayman’s Methyltransferase Colorimetric Assay Kit (Item No. 700140). Information represents the product specifications. Batch specific analytical results are provided on each certificate of analysis. Images SETD3 Acǎvity 1 2 3 250 kDa · · · · · · · 150 kDa · · · · · · · 100 kDa · · · · · · · 75 kDa · · · · · · · 50 kDa · · · · · · · 37 kDa · · · · · · · 25 kDa · · · · · · · 20 kDa · · · · · · · 15 kDa · · · · · · · Figure 2: Acǎvity Assay. SETD3 acǎvity was Lane 1: MW Markers determined using Cayman’s Methyltransferase Lane 2: SETD3 (2 µg) Colorimetric Assay Kit (Item No. 700140). Lane 3: SETD3 (4 µg) Figure 1: SDS-PAGE Analysis of SETD3 Representaìve gel image shown; actual purity may vary between each batch. WARNING CAYMAN CHEMICAL THIS PRODUCT IS FOR RESEARCH ONLY - NOT FOR HUMAN OR VETERINARY DIAGNOSTIC OR THERAPEUTIC USE. -

Biochemistry Entry of Fructose and Galactose
Paper : 04 Metabolism of carbohydrates Module : 06 Entry of Fructose and Galactose Dr. Vijaya Khader Dr. MC Varadaraj Principal Investigator Dr.S.K.Khare,Professor IIT Delhi. Paper Coordinator Dr. Ramesh Kothari,Professor UGC-CAS Department of Biosciences Saurashtra University, Rajkot-5, Gujarat-INDIA Dr. S. P. Singh, Professor Content Reviewer UGC-CAS Department of Biosciences Saurashtra University, Rajkot-5, Gujarat-INDIA Dr. Charmy Kothari, Assistant Professor Content Writer Department of Biotechnology Christ College, Affiliated to Saurashtra University, Rajkot-5, Gujarat-INDIA 1 Metabolism of Carbohydrates Biochemistry Entry of Fructose and Galactose Description of Module Subject Name Biochemistry Paper Name 04 Metabolism of Carbohydrates Module Name/Title 06 Entry of Fructose and Galactose 2 Metabolism of Carbohydrates Biochemistry Entry of Fructose and Galactose METABOLISM OF FRUCTOSE Objectives 1. To study the major pathway of fructose metabolism 2. To study specialized pathways of fructose metabolism 3. To study metabolism of galactose 4. To study disorders of galactose metabolism 3 Metabolism of Carbohydrates Biochemistry Entry of Fructose and Galactose Introduction Sucrose disaccharide contains glucose and fructose as monomers. Sucrose can be utilized as a major source of energy. Sucrose includes sugar beets, sugar cane, sorghum, maple sugar pineapple, ripe fruits and honey Corn syrup is recognized as high fructose corn syrup which gives the impression that it is very rich in fructose content but the difference between the fructose content in sucrose and high fructose corn syrup is only 5-10%. HFCS is rich in fructose because the sucrose extracted from the corn syrup is treated with the enzyme that converts some glucose in fructose which makes it more sweet. -

Supplementary Materials
Supplementary materials Supplementary Table S1: MGNC compound library Ingredien Molecule Caco- Mol ID MW AlogP OB (%) BBB DL FASA- HL t Name Name 2 shengdi MOL012254 campesterol 400.8 7.63 37.58 1.34 0.98 0.7 0.21 20.2 shengdi MOL000519 coniferin 314.4 3.16 31.11 0.42 -0.2 0.3 0.27 74.6 beta- shengdi MOL000359 414.8 8.08 36.91 1.32 0.99 0.8 0.23 20.2 sitosterol pachymic shengdi MOL000289 528.9 6.54 33.63 0.1 -0.6 0.8 0 9.27 acid Poricoic acid shengdi MOL000291 484.7 5.64 30.52 -0.08 -0.9 0.8 0 8.67 B Chrysanthem shengdi MOL004492 585 8.24 38.72 0.51 -1 0.6 0.3 17.5 axanthin 20- shengdi MOL011455 Hexadecano 418.6 1.91 32.7 -0.24 -0.4 0.7 0.29 104 ylingenol huanglian MOL001454 berberine 336.4 3.45 36.86 1.24 0.57 0.8 0.19 6.57 huanglian MOL013352 Obacunone 454.6 2.68 43.29 0.01 -0.4 0.8 0.31 -13 huanglian MOL002894 berberrubine 322.4 3.2 35.74 1.07 0.17 0.7 0.24 6.46 huanglian MOL002897 epiberberine 336.4 3.45 43.09 1.17 0.4 0.8 0.19 6.1 huanglian MOL002903 (R)-Canadine 339.4 3.4 55.37 1.04 0.57 0.8 0.2 6.41 huanglian MOL002904 Berlambine 351.4 2.49 36.68 0.97 0.17 0.8 0.28 7.33 Corchorosid huanglian MOL002907 404.6 1.34 105 -0.91 -1.3 0.8 0.29 6.68 e A_qt Magnogrand huanglian MOL000622 266.4 1.18 63.71 0.02 -0.2 0.2 0.3 3.17 iolide huanglian MOL000762 Palmidin A 510.5 4.52 35.36 -0.38 -1.5 0.7 0.39 33.2 huanglian MOL000785 palmatine 352.4 3.65 64.6 1.33 0.37 0.7 0.13 2.25 huanglian MOL000098 quercetin 302.3 1.5 46.43 0.05 -0.8 0.3 0.38 14.4 huanglian MOL001458 coptisine 320.3 3.25 30.67 1.21 0.32 0.9 0.26 9.33 huanglian MOL002668 Worenine -

(12) Patent Application Publication (10) Pub. No.: US 2003/0082511 A1 Brown Et Al
US 20030082511A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2003/0082511 A1 Brown et al. (43) Pub. Date: May 1, 2003 (54) IDENTIFICATION OF MODULATORY Publication Classification MOLECULES USING INDUCIBLE PROMOTERS (51) Int. Cl." ............................... C12O 1/00; C12O 1/68 (52) U.S. Cl. ..................................................... 435/4; 435/6 (76) Inventors: Steven J. Brown, San Diego, CA (US); Damien J. Dunnington, San Diego, CA (US); Imran Clark, San Diego, CA (57) ABSTRACT (US) Correspondence Address: Methods for identifying an ion channel modulator, a target David B. Waller & Associates membrane receptor modulator molecule, and other modula 5677 Oberlin Drive tory molecules are disclosed, as well as cells and vectors for Suit 214 use in those methods. A polynucleotide encoding target is San Diego, CA 92121 (US) provided in a cell under control of an inducible promoter, and candidate modulatory molecules are contacted with the (21) Appl. No.: 09/965,201 cell after induction of the promoter to ascertain whether a change in a measurable physiological parameter occurs as a (22) Filed: Sep. 25, 2001 result of the candidate modulatory molecule. Patent Application Publication May 1, 2003 Sheet 1 of 8 US 2003/0082511 A1 KCNC1 cDNA F.G. 1 Patent Application Publication May 1, 2003 Sheet 2 of 8 US 2003/0082511 A1 49 - -9 G C EH H EH N t R M h so as se W M M MP N FIG.2 Patent Application Publication May 1, 2003 Sheet 3 of 8 US 2003/0082511 A1 FG. 3 Patent Application Publication May 1, 2003 Sheet 4 of 8 US 2003/0082511 A1 KCNC1 ITREXCHO KC 150 mM KC 2000000 so 100 mM induced Uninduced Steady state O 100 200 300 400 500 600 700 Time (seconds) FIG. -

Characterization of Visceral and Subcutaneous Adipose Tissue
J. Perinat. Med. 2016; 44(7): 813–835 Shali Mazaki-Tovi*, Adi L. Tarca, Edi Vaisbuch, Juan Pedro Kusanovic, Nandor Gabor Than, Tinnakorn Chaiworapongsa, Zhong Dong, Sonia S. Hassan and Roberto Romero* Characterization of visceral and subcutaneous adipose tissue transcriptome in pregnant women with and without spontaneous labor at term: implication of alternative splicing in the metabolic adaptations of adipose tissue to parturition DOI 10.1515/jpm-2015-0259 groups (unpaired analyses) and adipose tissue regions Received July 27, 2015. Accepted October 26, 2015. Previously (paired analyses). Selected genes were tested by quantita- published online March 19, 2016. tive reverse transcription-polymerase chain reaction. Abstract Results: Four hundred and eighty-two genes were differ- entially expressed between visceral and subcutaneous Objective: The aim of this study was to determine gene fat of pregnant women with spontaneous labor at term expression and splicing changes associated with par- (q-value < 0.1; fold change > 1.5). Biological processes turition and regions (visceral vs. subcutaneous) of the enriched in this comparison included tissue and vascu- adipose tissue of pregnant women. lature development as well as inflammatory and meta- Study design: The transcriptome of visceral and abdomi- bolic pathways. Differential splicing was found for 42 nal subcutaneous adipose tissue from pregnant women at genes [q-value < 0.1; differences in Finding Isoforms using term with (n = 15) and without (n = 25) spontaneous labor Robust Multichip Analysis scores > 2] between adipose was profiled with the Affymetrix GeneChip Human Exon tissue regions of women not in labor. Differential exon 1.0 ST array. Overall gene expression changes and the dif- usage associated with parturition was found for three ferential exon usage rate were compared between patient genes (LIMS1, HSPA5, and GSTK1) in subcutaneous tissues. -

SETD3 Is an Actin Histidine Methyltransferase That Prevents Primary Dystocia
HHS Public Access Author manuscript Author ManuscriptAuthor Manuscript Author Nature. Manuscript Author Author manuscript; Manuscript Author available in PMC 2019 June 10. Published in final edited form as: Nature. 2019 January ; 565(7739): 372–376. doi:10.1038/s41586-018-0821-8. SETD3 is an Actin Histidine Methyltransferase that Prevents Primary Dystocia Alex W. Wilkinson1,*, Jonathan Diep2,*, Shaobo Dai3,*, Shuo Liu1, Yaw shin Ooi2, Dan Song4, Tie-Mei Li1, John R. Horton3, Xing Zhang3, Chao Liu4, Darshan V. Trivedi4, Katherine M. Ruppel4, José G. Vilches-Moure5, Kerriann M. Casey5, Justin Mak6, Tina Cowan7, Joshua E. Elias8, Claude M. Nagamine5, James A. Spudich4, Xiaodong Cheng3,#, Jan E. Carette2,#, and Or Gozani1,# 1Department of Biology, Stanford University, Stanford, CA 94305, USA 2Department of Microbiology and Immunology, Stanford University School of Medicine, Stanford, CA 94305, USA 3Department of Molecular and Cellular Oncology, The University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA 4Department of Biochemistry, Stanford University School of Medicine, Stanford, CA 94305, USA 5Department of Comparative Medicine, Stanford University School of Medicine, Stanford, CA 94305, USA 6Stanford Healthcare, Palo Alto, CA 94305 USA 7Departent of Pathology, Stanford University School of Medicine, Stanford, CA 94305, USA 8Deparment of Chemical and Systems Biology, Stanford University School of Medicine, Stanford CA 94305, USA Abstract For over fifty years, the methylation of mammalian actin at histidine 73 (actin-H73me) has been known to exist1. Beyond mammals, we find that actin-H73me is conserved in several additional model animal and plant organisms. Despite the pervasiveness of H73me, its function is enigmatic, and the enzyme generating this modification is unknown. -

Lipoprotein Lipase: a General Review Moacir Couto De Andrade Júnior1,2*
Review Article iMedPub Journals Insights in Enzyme Research 2018 www.imedpub.com Vol.2 No.1:3 ISSN 2573-4466 DOI: 10.21767/2573-4466.100013 Lipoprotein Lipase: A General Review Moacir Couto de Andrade Júnior1,2* 1Post-Graduation Department, Nilton Lins University, Manaus, Amazonas, Brazil 2Department of Food Technology, Instituto Nacional de Pesquisas da Amazônia (INPA), Manaus, Amazonas, Brazil *Corresponding author: MC Andrade Jr, Post-Graduation Department, Nilton Lins University, Manaus, Amazonas, Brazil, Tel: +55 (92) 3633-8028; E-mail: [email protected] Rec date: March 07, 2018; Acc date: April 10, 2018; Pub date: April 17, 2018 Copyright: © 2018 Andrade Jr MC. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Citation: Andrade Jr MC (2018) Lipoprotein Lipase: A General Review. Insights Enzyme Res Vol.2 No.1:3 Abstract Lipoprotein Lipase: Historical Hallmarks, Enzymatic Activity, Characterization, and Carbohydrates (e.g., glucose) and lipids (e.g., free fatty acids or FFAs) are the most important sources of energy Present Relevance in Human for most organisms, including humans. Lipoprotein lipase (LPL) is an extracellular enzyme (EC 3.1.1.34) that is Pathophysiology and Therapeutics essential in lipoprotein metabolism. LPL is a glycoprotein that is synthesized and secreted in several tissues (e.g., Macheboeuf, in 1929, first described chemical procedures adipose tissue, skeletal muscle, cardiac muscle, and for the isolation of a plasma protein fraction that was very rich macrophages). At the luminal surface of the vascular in lipids but readily soluble in water, such as a lipoprotein [1]. -

Genetic Alterations of Histone Lysine Methyltransferases and Their Significance in Breast Cancer
www.impactjournals.com/oncotarget/ Oncotarget, Vol. 6, No.4 Genetic alterations of histone lysine methyltransferases and their significance in breast cancer Lanxin Liu1,*, Sarah Kimball1,*, Hui Liu1, Andreana Holowatyj1 and Zeng-Quan Yang1 1 Department of Oncology, Karmanos Cancer Institute, Wayne State University, Detroit, MI, USA * These authors contributed equally to this work Correspondence to: Zeng-Quan Yang, email: [email protected] Keywords: breast cancer, histone lysine methyltransferase, gene amplification, deletion, mutation Received: August 27, 2014 Accepted: December 10, 2014 Published: December 11, 2014 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT Histone lysine methyltransferases (HMTs), a large class of enzymes that catalyze site-specific methylation of lysine residues on histones and other proteins, play critical roles in controlling transcription, chromatin architecture, and cellular differentiation. However, the genomic landscape and clinical significance of HMTs in breast cancer remain poorly characterized. Here, we conducted a meta-analysis of approximately 50 HMTs in breast cancer and identified associations among recurrent copy number alterations, mutations, gene expression, and clinical outcome. We identified 12 HMTs with the highest frequency of genetic alterations, including 8 with high-level amplification, 2 with putative homozygous deletion, and 2 with somatic mutation. Different subtypes of breast cancer have different patterns of copy number and expression for each HMT gene. In addition, chromosome 1q contains four HMTs that are concurrently or independently amplified or overexpressed in breast cancer. Copy number or mRNA expression of several HMTs was significantly associated with basal- like breast cancer and shorter patient survival. -

Properties and Kinetic Analysis of UDP-Glucose Dehydrogenase from Group a Streptococci IRREVERSIBLE INHIBITION by UDP-CHLOROACETOL*
THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 272, No. 6, Issue of February 7, pp. 3416–3422, 1997 © 1997 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. Properties and Kinetic Analysis of UDP-glucose Dehydrogenase from Group A Streptococci IRREVERSIBLE INHIBITION BY UDP-CHLOROACETOL* (Received for publication, September 19, 1996, and in revised form, November 6, 1996) Robert E. Campbell‡§, Rafael F. Sala‡, Ivo van de Rijn¶, and Martin E. Tanner‡i From the ‡Department of Chemistry, University of British Columbia, Vancouver, British Columbia V6T 1Z1, Canada and ¶Wake Forest University Medical Center, Winston-Salem, North Carolina 27157 UDP-glucuronic acid is used by many pathogenic bac- the capsule enables the bacteria to evade the host’s immune teria in the construction of an antiphagocytic capsule system (7, 8). Group A and C streptococci are mammalian that is required for virulence. The enzyme UDP-glucose pathogens that use UDPGDH in the synthesis of a capsule dehydrogenase catalyzes the NAD1-dependent 2-fold ox- composed of hyaluronic acid (a polysaccharide consisting of idation of UDP-glucose and provides a source of the alternating glucuronic acid and N-acetylglucosamine residues) acid. In the present study the recombinant dehydrogen- (9, 10). Many of the known strains of Streptococcus pneumoniae ase from group A streptococci has been purified and also use UDP-glucuronic acid in the construction of their po- found to be active as a monomer. The enzyme contains lysaccharide capsule (11), and it has recently been shown that no chromophoric cofactors, and its activity is unaffected UDPGDH is required for capsule production in S. -

Endogenous Protein Interactome of Human UDP-Glucuronosyltransferases Exposed by Untargeted Proteomics
ORIGINAL RESEARCH published: 03 February 2017 doi: 10.3389/fphar.2017.00023 Endogenous Protein Interactome of Human UDP-Glucuronosyltransferases Exposed by Untargeted Proteomics Michèle Rouleau, Yannick Audet-Delage, Sylvie Desjardins, Mélanie Rouleau, Camille Girard-Bock and Chantal Guillemette * Pharmacogenomics Laboratory, Canada Research Chair in Pharmacogenomics, Faculty of Pharmacy, Centre Hospitalier Universitaire de Québec Research Center, Laval University, Québec, QC, Canada The conjugative metabolism mediated by UDP-glucuronosyltransferase enzymes (UGTs) significantly influences the bioavailability and biological responses of endogenous molecule substrates and xenobiotics including drugs. UGTs participate in the regulation of cellular homeostasis by limiting stress induced by toxic molecules, and by Edited by: controlling hormonal signaling networks. Glucuronidation is highly regulated at genomic, Yuji Ishii, transcriptional, post-transcriptional and post-translational levels. However, the UGT Kyushu University, Japan protein interaction network, which is likely to influence glucuronidation, has received Reviewed by: little attention. We investigated the endogenous protein interactome of human UGT1A Ben Lewis, Flinders University, Australia enzymes in main drug metabolizing non-malignant tissues where UGT expression is Shinichi Ikushiro, most prevalent, using an unbiased proteomics approach. Mass spectrometry analysis Toyama Prefectural University, Japan of affinity-purified UGT1A enzymes and associated protein complexes in liver,