Hynobius Retardatus)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

50 CFR Ch. I (10–1–20 Edition) § 16.14
§ 15.41 50 CFR Ch. I (10–1–20 Edition) Species Common name Serinus canaria ............................................................. Common Canary. 1 Note: Permits are still required for this species under part 17 of this chapter. (b) Non-captive-bred species. The list 16.14 Importation of live or dead amphib- in this paragraph includes species of ians or their eggs. non-captive-bred exotic birds and coun- 16.15 Importation of live reptiles or their tries for which importation into the eggs. United States is not prohibited by sec- Subpart C—Permits tion 15.11. The species are grouped tax- onomically by order, and may only be 16.22 Injurious wildlife permits. imported from the approved country, except as provided under a permit Subpart D—Additional Exemptions issued pursuant to subpart C of this 16.32 Importation by Federal agencies. part. 16.33 Importation of natural-history speci- [59 FR 62262, Dec. 2, 1994, as amended at 61 mens. FR 2093, Jan. 24, 1996; 82 FR 16540, Apr. 5, AUTHORITY: 18 U.S.C. 42. 2017] SOURCE: 39 FR 1169, Jan. 4, 1974, unless oth- erwise noted. Subpart E—Qualifying Facilities Breeding Exotic Birds in Captivity Subpart A—Introduction § 15.41 Criteria for including facilities as qualifying for imports. [Re- § 16.1 Purpose of regulations. served] The regulations contained in this part implement the Lacey Act (18 § 15.42 List of foreign qualifying breed- U.S.C. 42). ing facilities. [Reserved] § 16.2 Scope of regulations. Subpart F—List of Prohibited Spe- The provisions of this part are in ad- cies Not Listed in the Appen- dition to, and are not in lieu of, other dices to the Convention regulations of this subchapter B which may require a permit or prescribe addi- § 15.51 Criteria for including species tional restrictions or conditions for the and countries in the prohibited list. -
I Online Supplementary Data – Sexual Size Dimorphism in Salamanders
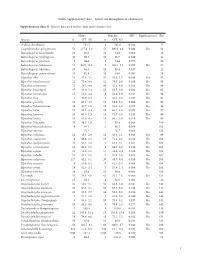
Online Supplementary data – Sexual size dimorphism in salamanders Supplementary data S1. Species data used in this study and references list. Males Females SSD Significant test Ref Species n SVL±SD n SVL±SD Andrias davidianus 2 532.5 8 383.0 -0.280 12 Cryptobranchus alleganiensis 53 277.4±5.2 52 300.9±3.4 0.084 Yes 61 Batrachuperus karlschmidti 10 80.0 10 84.8 0.060 26 Batrachuperus londongensis 20 98.6 10 96.7 -0.019 12 Batrachuperus pinchonii 5 69.6 5 74.6 0.070 26 Batrachuperus taibaiensis 11 92.9±12.1 9 102.1±7.1 0.099 Yes 27 Batrachuperus tibetanus 10 94.5 10 92.8 -0.017 12 Batrachuperus yenyuadensis 10 82.8 10 74.8 -0.096 26 Hynobius abei 24 57.8±2.1 34 55.0±1.2 -0.048 Yes 92 Hynobius amakusaensis 22 75.4±4.8 12 76.5±3.6 0.014 No 93 Hynobius arisanensis 72 54.3±4.8 40 55.2±4.8 0.016 No 94 Hynobius boulengeri 37 83.0±5.4 15 91.5±3.8 0.102 Yes 95 Hynobius formosanus 15 53.0±4.4 8 52.4±3.9 -0.011 No 94 Hynobius fuca 4 50.9±2.8 3 52.8±2.0 0.037 No 94 Hynobius glacialis 12 63.1±4.7 11 58.9±5.2 -0.066 No 94 Hynobius hidamontanus 39 47.7±1.0 15 51.3±1.2 0.075 Yes 96 Hynobius katoi 12 58.4±3.3 10 62.7±1.6 0.073 Yes 97 Hynobius kimurae 20 63.0±1.5 15 72.7±2.0 0.153 Yes 98 Hynobius leechii 70 61.6±4.5 18 66.5±5.9 0.079 Yes 99 Hynobius lichenatus 37 58.5±1.9 2 53.8 -0.080 100 Hynobius maoershanensis 4 86.1 2 80.1 -0.069 101 Hynobius naevius 72.1 76.7 0.063 102 Hynobius nebulosus 14 48.3±2.9 12 50.4±2.1 0.043 Yes 96 Hynobius osumiensis 9 68.4±3.1 15 70.2±3.0 0.026 No 103 Hynobius quelpaertensis 41 52.5±3.8 4 61.3±4.1 0.167 Yes 104 Hynobius -

Salamander Species Listed As Injurious Wildlife Under 50 CFR 16.14 Due to Risk of Salamander Chytrid Fungus Effective January 28, 2016
Salamander Species Listed as Injurious Wildlife Under 50 CFR 16.14 Due to Risk of Salamander Chytrid Fungus Effective January 28, 2016 Effective January 28, 2016, both importation into the United States and interstate transportation between States, the District of Columbia, the Commonwealth of Puerto Rico, or any territory or possession of the United States of any live or dead specimen, including parts, of these 20 genera of salamanders are prohibited, except by permit for zoological, educational, medical, or scientific purposes (in accordance with permit conditions) or by Federal agencies without a permit solely for their own use. This action is necessary to protect the interests of wildlife and wildlife resources from the introduction, establishment, and spread of the chytrid fungus Batrachochytrium salamandrivorans into ecosystems of the United States. The listing includes all species in these 20 genera: Chioglossa, Cynops, Euproctus, Hydromantes, Hynobius, Ichthyosaura, Lissotriton, Neurergus, Notophthalmus, Onychodactylus, Paramesotriton, Plethodon, Pleurodeles, Salamandra, Salamandrella, Salamandrina, Siren, Taricha, Triturus, and Tylototriton The species are: (1) Chioglossa lusitanica (golden striped salamander). (2) Cynops chenggongensis (Chenggong fire-bellied newt). (3) Cynops cyanurus (blue-tailed fire-bellied newt). (4) Cynops ensicauda (sword-tailed newt). (5) Cynops fudingensis (Fuding fire-bellied newt). (6) Cynops glaucus (bluish grey newt, Huilan Rongyuan). (7) Cynops orientalis (Oriental fire belly newt, Oriental fire-bellied newt). (8) Cynops orphicus (no common name). (9) Cynops pyrrhogaster (Japanese newt, Japanese fire-bellied newt). (10) Cynops wolterstorffi (Kunming Lake newt). (11) Euproctus montanus (Corsican brook salamander). (12) Euproctus platycephalus (Sardinian brook salamander). (13) Hydromantes ambrosii (Ambrosi salamander). (14) Hydromantes brunus (limestone salamander). (15) Hydromantes flavus (Mount Albo cave salamander). -

Title 50—Wildlife and Fisheries
Title 50—Wildlife and Fisheries (This book contains parts 1 to 16) Part CHAPTER I—United States Fish and Wildlife Service, Depart- ment of the Interior ........................................................... 1 1 VerDate Sep<11>2014 11:47 Dec 15, 2016 Jkt 238234 PO 00000 Frm 00011 Fmt 8008 Sfmt 8008 Y:\SGML\238234.XXX 238234 jstallworth on DSK7TPTVN1PROD with CFR VerDate Sep<11>2014 11:47 Dec 15, 2016 Jkt 238234 PO 00000 Frm 00012 Fmt 8008 Sfmt 8008 Y:\SGML\238234.XXX 238234 jstallworth on DSK7TPTVN1PROD with CFR CHAPTER I—UNITED STATES FISH AND WILDLIFE SERVICE, DEPARTMENT OF THE INTERIOR SUBCHAPTER A—GENERAL PROVISIONS Part Page 1 Definitions .............................................................. 5 2 Agency organization and locations ......................... 5 3 Nondiscrimination—contracts, permits, and use of facilities ............................................................... 7 SUBCHAPTER B—TAKING, POSSESSION, TRANSPORTATION, SALE, PUR- CHASE, BARTER, EXPORTATION, AND IMPORTATION OF WILDLIFE AND PLANTS 10 General provisions.................................................. 8 11 Civil procedures...................................................... 30 12 Seizure and forfeiture procedures ........................... 34 13 General permit procedures ...................................... 44 14 Importation, exportation, and transportation of wildlife ................................................................. 57 15 Wild Bird Conservation Act .................................... 85 16 Injurious wildlife..................................................... 98 3 VerDate Sep<11>2014 11:47 Dec 15, 2016 Jkt 238234 PO 00000 Frm 00013 Fmt 8008 Sfmt 8008 Y:\SGML\238234.XXX 238234 jstallworth on DSK7TPTVN1PROD with CFR VerDate Sep<11>2014 11:47 Dec 15, 2016 Jkt 238234 PO 00000 Frm 00014 Fmt 8008 Sfmt 8008 Y:\SGML\238234.XXX 238234 jstallworth on DSK7TPTVN1PROD with CFR SUBCHAPTER A—GENERAL PROVISIONS PART 1—DEFINITIONS § 1.6 Person. Person means an individual, club, as- Sec. sociation, partnership, corporation, or 1.1 Meaning of terms. -

Herpetology~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~Of11 Japan
OF THE HERPETOLOGY~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~OF11 JAPAN CARL GANS BULLETIN. OF THE AMERIC,AN MUSEUM OF NATU-RAL. HISTORY VOLUME ~~93:~ARTICLE 6 NEW'YORK.:'l949NWYR:14 M-- A BIBLIOGRAPHY OF THE HERPETOLOGY OF JAPAN A BIBLIOGRAPHY OF THE HERPETOLOGY OF JAPAN CARL GANS BULLETIN OF THE AMERICAN MUSEUM OF NATURAL HISTORY VOLUME 93 :ARTICLE 6 NEW YORK:1949 BULLETIN OF THE AMERICAN MUSEUM OF NATURAL HISTORY Volume 93, article 6, pages 389-496 Issued July 12, 1949 Price: $1.25 a copy INTRODUCTION THROUGHOUT HISTORY one of the chief prob- History. A supplement to, or perhaps a re- lems facing investigators in the natural sci- vised edition of, this Bibliography will be ences has been the communication of their published if the response warrants it. ideas and the results of their work to fellow investigators. The main difficulty in this re- SCOPE spect may date back to Biblical times when The papers included have been confined to at Babel "the Lord did there confound the those dealing with recent reptiles and am- language of the earth."1 Much duplication of phibians of the chain of islands lying off the research has resulted from the fact that rec- coast of Asia, enumerated as follows: the four ords of earlier work in other countries have main islands of Japan (Honshu, Hokkaido, been unavailable to investigators. Kyushu, and Shikoku), the Kurile, Bonin, Recognition of the fact that this condition and Riu Kiu archipelagoes, and the islands existed in regard to the fauna of Japan was of Tsushima and Formosa. For purposes of forcibly brought to my attention during the comparison a few papers dealing with the course of a year's stay in that archipelago. -

Title Systematic Studies of Two Japanese Brown Frogs
Systematic studies of two Japanese brown frogs( Title Dissertation_全文 ) Author(s) Eto, Koshiro Citation Kyoto University (京都大学) Issue Date 2014-03-24 URL http://dx.doi.org/10.14989/doctor.k18358 Right 許諾条件により本文は2015-03-23に公開 Type Thesis or Dissertation Textversion ETD Kyoto University Systematic studies of two Japanese brown frogs Koshiro ETO 2013 A calling male of Tago’s brown frog, Rana tagoi (above) and an amplectant pair of Stream brown frog, R. sakuraii (below). CONTENTS Page Contents Chapter 1 Introduction 1 Chapter 2 Highly Complex Mitochondrial DNA Genealogy in an 4 Endemic Japanese Subterranean Breeding Brown Frog Rana tagoi (Amphibia, Anura, Ranidae) Chapter 3 Discordance between Mitochondrial DNA Genealogy and 27 Nuclear DNA Genetic Structure in the Two Morphotypes of Rana tagoi tagoi in the Kinki Region, Japan Chapter 4 Cytonuclear Discordance and Historical Demography of Two 42 Brown Frogs Rana tagoi and R. sakuraii Chapter 5 General Discussion 75 Summary 79 Acknowledgements 80 References 81 Appendix 91 i CHAPTER 1 Introduction As is well known, Mayr et al . (1953) recognized three levels (alpha, beta, and gamma) in the study of taxonomy, although they are mutually correlated and are not clear-cut. For Japanese amphibians, the alpha-level taxonomic studies started in the middle of 19 th century (Temminck & Schlegel 1838), and through the later studies by authors like Stejneger (1907), Okada (1930), and Nakamura & Ueno (1963), basic species classification was established by the 1960s. Then the introduction of modern methodologies like molecular analyses enabled studies of relationships among species, i.e., beta taxonomy (Matsui 2000). Studies using allozymic techniques started over 30 years ago, and resulted in many findings related to taxonomy (e.g., Matsui & Miyazaki 1984; Matsui 1987, 1994; Matsui et al. -

DEPARTMENT of the INTERIOR Fish and Wildlife
This document is scheduled to be published in the Federal Register on 01/13/2016 and available online at http://federalregister.gov/a/2016-00452, and on FDsys.gov DEPARTMENT OF THE INTERIOR Fish and Wildlife Service 50 CFR Part 16 RIN 1018–BA77 [Docket No. FWS–HQ–FAC–2015–0005] [FXFR13360900000–156–FF09F14000] Injurious Wildlife Species; Listing Salamanders Due to Risk of Salamander Chytrid Fungus AGENCY: Fish and Wildlife Service, Interior. ACTION: Interim rule; request for comments; notice of availability of economic analysis. 1 SUMMARY: The U.S. Fish and Wildlife Service is amending its regulations under the Lacey Act to add all species of salamanders from 20 genera, of which there are 201 species, to the list of injurious amphibians. With this interim rule, both importation into the United States and interstate transportation between States, the District of Columbia, the Commonwealth of Puerto Rico, or any territory or possession of the United States of any live or dead specimen, including parts, of these 20 genera of salamanders are prohibited, except by permit for zoological, educational, medical, or scientific purposes (in accordance with permit conditions) or by Federal agencies without a permit solely for their own use. This action is necessary to protect the interests of wildlife and wildlife resources from the introduction, establishment, and spread of the chytrid fungus Batrachochytrium salamandrivorans into ecosystems of the United States. The fungus affects salamanders, with lethal effects on many species, and is not yet known to be found in the United States. Because of the devastating effect that we expect the fungus will have on native U.S. -

Hynobius Nebulosus)
A Skeletochronological Study of Breeding Females in a Title Population of Japanese Clouded Salamanders (Hynobius nebulosus) Author(s) Matsuki, Takashi; Matsui, Masafumi Citation Zoological Science (2011), 28(3): 175-179 Issue Date 2011-03 URL http://hdl.handle.net/2433/216895 Right © 2011 Zoological Society of Japan Type Journal Article Textversion publisher Kyoto University ZOOLOGICAL SCIENCE 28: 175–179 (2011) ¤ 2011 Zoological Society of Japan A Skeletochronological Study of Breeding Females in a Population of Japanese Clouded Salamanders (Hynobius nebulosus) Takashi Matsuki and Masafumi Matsui* Graduate School of Human and Environmental Studies, Kyoto University, Yoshida Nihonmatsu-cho, Sakyo-ku, Kyoto 606-8501, Japan The age structure of breeding females of Hynobius nebulosus has not been studied sufficiently. We estimated the ages of 76 individuals from a population in Kyoto by using skeletochronology. The mean age and snout-vent length (SVL) of this population were 4.6 years and 55.7 mm, respectively. It was estimated that the youngest females breed two years post hatching at a mean SVL of 46.5 mm, but a larger number of individuals begins breeding at three years and a mean SVL of 52.2 mm. Because most males also start to breed at three years, there seems to be no gender difference in the timing of sexual maturation. The age of the oldest female was estimated to be 11.8 years. It is possible that the life history of H. nebulosus is characterized by early maturation and arrested growth, and short longevity. Key words: Life history traits, skeletochronology, age structure, age at first reproduction, longevity, Hynobius nebulosus ficulty was overcome by introduction of the drift fence trap. -

JSSZ: a Journal "Proceedings ..."
http://wwwsoc.nii.ac.jp/jssz2/proc/index.html [English top page]/[Japanese top page] Report of the Japanese Society of Systematic Zoology, No. 1 (1965) Proceeding of the Japanese Society of Systematic Zoology, Nos. 2-6 (1966-1970) Proceedings of the Japanese Society of Systematic Zoology, Nos. 7-54 (1971-1995) (ISSN 0287-0223) Proceedings of the Japanese Society of Systematic Zoology is a previous journal of The Society. Back issues are available. Ordinary Issues: Nos. 1-22, 24-26, 28-31, 33-42, 44, 45, 47-54. ● Contents ordered by author name: ❍ A, B, C, D, E, F, G, H, I, J, K, L, M, N, O, P, Q, R, S, T, U, V, W, X, Y, Z ● Contents ordered by subject: ❍ theory; animals in general; other animals; othre articles ❍ protozoans ❍ Porifera; Cnidaria ❍ Plathelminthes; Nemertinea; Kamptozoa ❍ Aschelminthes; Mollusca ❍ Annelida; Tardigrada ❍ Arthropoda, Xiphosura ❍ Arthropoda, Aracnida ❍ Arthropoda, Crustacea ❍ Arthropoda, myriapodans ❍ Arthropoda, Insecta ❍ Bryozoa; Chaetognatha; Echinodermata; Chordata Special Issues: No. 23 No. 27 No. 32 No. 43: The Rotifera from Singapore and Taiwan. No. 46: Taxonomical and Ecological Approaches to the Aquatic Biota in the Southwestern Islands of Japan. Report of the Japanese Society of Systematic Zoology: No. 1 (Sept 10, 1965), Proceeding of the Japanese Society of Systematic Zoology: No. 2 (Aug 30, 1966), No. 3 (Sept 10, 1967), No. 4 (Oct 1, 1968), No. 5 (Oct 1, 1969), No. 6 (Oct 1, 1970), Proceedings of the Japanese Society of Systematic Zoology: No. 7 (Oct 1, 1971), No. 8 (Nov 15, 1972), No. 9 (Oct 20, 1973), No. 10 (Dec 14, 1974), No. -

Federal Register/Vol. 81, No. 8/Wednesday, January 13, 2016
1534 Federal Register / Vol. 81, No. 8 / Wednesday, January 13, 2016 / Rules and Regulations If . Then include . ******* (j) the estimated value of the acquisition exceeds $10 million ................ 52.222–24 Pre-award On-site Equal Opportunity Compliance Evalua- tion. (k) the contracting officer requires cost or pricing data for work or serv- 52.215–10 Price Reduction for Defective Certified Cost or Pricing Data. ices exceeding the threshold identified in FAR 15.403–4. 52.215–12 Subcontractor Certified Cost or Pricing Data. ******* [FR Doc. 2016–00475 Filed 1–12–16; 8:45 am] DATES: This interim rule is effective as the offspring or eggs of any of the BILLING CODE 6820–161–P of January 28, 2016. Interested persons foregoing that are injurious to human are invited to submit written comments beings, to the interests of agriculture, on this interim rule on or before March horticulture, or forestry, or to the DEPARTMENT OF THE INTERIOR 14, 2016 wildlife or wildlife resources of the ADDRESSES: You may submit comments United States. Fish and Wildlife Service by any of the following methods: We have determined that salamanders • Federal eRulemaking Portal: http:// that can carry the fungus 50 CFR Part 16 www.regulations.gov. Search for Docket Batrachochytrium salamandrivorans (Bsal) are injurious to wildlife and RIN 1018–BA77 No. FWS–HQ–FAC–2015–0005 and follow the instructions for submitting wildlife resources of the United States. [Docket No. FWS–HQ–FAC–2015–0005; comments. This determination was based on a FXFR13360900000–156–FF09F14000] • Mail, Hand Delivery, or Courier: review of the literature and an Public Comments Processing, Attn: evaluation under the criteria for Injurious Wildlife Species; Listing FWS–HQ–FAC–2015–0005; Division of injuriousness by the Service. -

A Thesis Submitted to the Honors College in Partial Fulfillment of the Bachelors Degree with Honors in Natural Resources
The Biogeography Of Diel Patterns In Herpetofauna Item Type text; Electronic Thesis Authors Runnion, Emily Publisher The University of Arizona. Rights Copyright © is held by the author. Digital access to this material is made possible by the University Libraries, University of Arizona. Further transmission, reproduction or presentation (such as public display or performance) of protected items is prohibited except with permission of the author. Download date 27/09/2021 09:59:20 Item License http://rightsstatements.org/vocab/InC/1.0/ Link to Item http://hdl.handle.net/10150/632848 THE BIOGEOGRAPHY OF DIEL PATTERNS IN HERPETOFAUNA By EMILY RUNNION _______________________________ A Thesis Submitted to The Honors College In Partial Fulfillment of the Bachelors degree With Honors in Natural Resources THE UNIVERSITY OF ARIZONA MAY 2019 Approved by: _____________________________ Dr. John Wiens Ecology and Evolutionary Biology ABSTRACT Animal activity varies greatly during daylight and nighttime hours, with species ranging from strictly diurnal or nocturnal, to cathemeral or crepuscular. Which of these categories a species falls into may be determined by a variety of factors, including temperature, and biogeographic location. In this study I analyzed diel activity patterns at 13 sites distributed around the world, encompassing a total of 901 species of reptile and amphibians. I compared these activity patterns to the absolute latitudes of each site and modeled diel behavior as a function of distance from the equator. I found that latitude does correlate with the presence of nocturnal snake and lizard species. However, I did not find significant results correlating latitude to any other reptile or amphibian lineage. INTRODUCTION Cycles of light and darkness are important for the activity and behaviors of many organisms, both eukaryotic and prokaryotic (Johnson et al. -

Population Structure of the Salamander Hynobius Retardatus Inferred from a Partial Sequence of the Mitochondrial DNA Title Control Region
Population structure of the salamander Hynobius retardatus inferred from a partial sequence of the mitochondrial DNA Title control region Author(s) Azuma, Noriko; Hangui, Jun-ichi; Wakahara, Masami; Michimae, Hirofumi Zoological science, 30(1), 7-14 Citation https://doi.org/10.2108/zsj.30.7 Issue Date 2013-01 Doc URL http://hdl.handle.net/2115/54544 Type article File Information 1)zsj.30.7[1].pdf Instructions for use Hokkaido University Collection of Scholarly and Academic Papers : HUSCAP Population Structure of the Salamander Hynobius retardatus Inferred from a Partial Sequence of the Mitochondrial DNA Control Region Author(s): Noriko Azuma , Jun-ichi Hangui , Masami Wakahara and Hirofumi Michimae Source: Zoological Science, 30(1):7-14. 2013. Published By: Zoological Society of Japan DOI: http://dx.doi.org/10.2108/zsj.30.7 URL: http://www.bioone.org/doi/full/10.2108/zsj.30.7 BioOne (www.bioone.org) is a nonprofit, online aggregation of core research in the biological, ecological, and environmental sciences. BioOne provides a sustainable online platform for over 170 journals and books published by nonprofit societies, associations, museums, institutions, and presses. Your use of this PDF, the BioOne Web site, and all posted and associated content indicates your acceptance of BioOne’s Terms of Use, available at www.bioone.org/page/terms_of_use. Usage of BioOne content is strictly limited to personal, educational, and non-commercial use. Commercial inquiries or rights and permissions requests should be directed to the individual publisher as copyright holder. BioOne sees sustainable scholarly publishing as an inherently collaborative enterprise connecting authors, nonprofit publishers, academic institutions, research libraries, and research funders in the common goal of maximizing access to critical research.