Gene Expression Evidence for Remodeling of Lateral Hypothalamic Circuitry in Cocaine Addiction
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Crispra Screening with Real World Evidence Identifies Potassium Channels As Neuronal Entry Factors and Druggable Targets for SARS-Cov-2
bioRxiv preprint doi: https://doi.org/10.1101/2021.07.01.450475; this version posted July 1, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. CRISPRa screening with real world evidence identifies potassium channels as neuronal entry factors and druggable targets for SARS-CoV-2 Authors: Chengkun Wang1,†, Ravi K. Dinesh1,†,*, Yuanhao Qu1,2,3,†, Arjun Rustagi4,†, Henry Cousins1,5,‡, James Zengel6,‡, Yinglong Guo7,‡, Taryn Hall7,‡, Aimee Beck4, Luke Tso7, EliF Tokar ErdemiC7, Kae Tanudtanud7, Sheng Ren7, Kathy Tzy-Hwa Tzeng7, Aaron Wilk4,5, Mengdi Wang8, Jan Carette2,6, Russ Altman2,4,9,*, Catherine A. Blish4,5,10,*, Le Cong1,2,3,* Affiliations: 1Department oF Pathology, StanFord University School oF Medicine, StanFord, CA, USA 2Department oF Genetics, StanFord University School oF Medicine, StanFord, CA, USA 3Cancer Biology Program, StanFord University School oF Medicine, StanFord, CA, USA 4Department oF Medicine, StanFord University School oF Medicine, StanFord, CA, USA 5Medical Scientist Training Program, StanFord University School oF Medicine, StanFord, CA, USA 6Department oF Microbiology and Immunology, StanFord University School oF Medicine, StanFord, CA, USA 7Research and Development at UnitedHealth Group, Minneapolis, MN, USA 8Center For Statistics and Machine Learning, Department oF Electrical and Computer Engineering, Princeton University, Princeton, NJ, USA 9Department oF Bioengineering, StanFord University, StanFord, CA, USA 10Chan Zuckerberg Biohub, San Francisco, CA, USA *Correspondence to: [email protected] (R.K.D.); [email protected] (R.A.); [email protected] (C.A.B.); [email protected] (L.C.) † These authors contributed equally to this work. -

Potassium Channels in Epilepsy
Downloaded from http://perspectivesinmedicine.cshlp.org/ on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press Potassium Channels in Epilepsy Ru¨diger Ko¨hling and Jakob Wolfart Oscar Langendorff Institute of Physiology, University of Rostock, Rostock 18057, Germany Correspondence: [email protected] This review attempts to give a concise and up-to-date overview on the role of potassium channels in epilepsies. Their role can be defined from a genetic perspective, focusing on variants and de novo mutations identified in genetic studies or animal models with targeted, specific mutations in genes coding for a member of the large potassium channel family. In these genetic studies, a demonstrated functional link to hyperexcitability often remains elusive. However, their role can also be defined from a functional perspective, based on dy- namic, aggravating, or adaptive transcriptional and posttranslational alterations. In these cases, it often remains elusive whether the alteration is causal or merely incidental. With 80 potassium channel types, of which 10% are known to be associated with epilepsies (in humans) or a seizure phenotype (in animals), if genetically mutated, a comprehensive review is a challenging endeavor. This goal may seem all the more ambitious once the data on posttranslational alterations, found both in human tissue from epilepsy patients and in chronic or acute animal models, are included. We therefore summarize the literature, and expand only on key findings, particularly regarding functional alterations found in patient brain tissue and chronic animal models. INTRODUCTION TO POTASSIUM evolutionary appearance of voltage-gated so- CHANNELS dium (Nav)andcalcium (Cav)channels, Kchan- nels are further diversified in relation to their otassium (K) channels are related to epilepsy newer function, namely, keeping neuronal exci- Psyndromes on many different levels, ranging tation within limits (Anderson and Greenberg from direct control of neuronal excitability and 2001; Hille 2001). -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Changes in Excitability and Ion Channel Expression in Neurons of the Major 2 Pelvic Ganglion in Female Type II Diabetic Mice
bioRxiv preprint doi: https://doi.org/10.1101/360826; this version posted July 4, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Changes in excitability and ion channel expression in neurons of the major 2 pelvic ganglion in female type II diabetic mice 3 4 5 Michael Gray1*, Kawasi M. Lett1*, Virginia B. Garcia1, Cindy Kyi1, Kathleen A. Pennington2, Laura C. 6 Schulz2, David J. Schulz1 7 8 1 Division of Biological Sciences, University of Missouri, Columbia, MO USA 65211 9 2 Department of Obstetrics, Gynecology and Women's Health, University of Missouri, Columbia, MO, 10 65211, USA. 11 * Denotes equal contribution by these authors 12 13 14 15 Abbreviated Title 16 Changes in parasympathetic neurons in Type II diabetes 17 18 Corresponding Author 19 David J. Schulz, Ph.D. 20 Division of Biological Sciences 21 University of Missouri-Columbia 22 218 LeFevre Hall 23 Columbia, MO 65211 24 Ph 573-882-4067 25 Fax 573-884-5020 26 Email: [email protected] 27 28 29 Acknowledgments 30 This work was funded by a grant from the Missouri Spinal Cord Injuries Research Program (D.J.S.), the 31 Craig H. Neilsen Foundation (D.J.S.), American Diabetes Association Grant 1-14-BS-181 (L.C.S.) and 32 American Heart Association Postdoctoral Fellowship 13POST16910108 (K.A.P.). The authors declare no 33 competing financial interests. -

Ion Channels 3 1
r r r Cell Signalling Biology Michael J. Berridge Module 3 Ion Channels 3 1 Module 3 Ion Channels Synopsis Ion channels have two main signalling functions: either they can generate second messengers or they can function as effectors by responding to such messengers. Their role in signal generation is mainly centred on the Ca2 + signalling pathway, which has a large number of Ca2+ entry channels and internal Ca2+ release channels, both of which contribute to the generation of Ca2 + signals. Ion channels are also important effectors in that they mediate the action of different intracellular signalling pathways. There are a large number of K+ channels and many of these function in different + aspects of cell signalling. The voltage-dependent K (KV) channels regulate membrane potential and + excitability. The inward rectifier K (Kir) channel family has a number of important groups of channels + + such as the G protein-gated inward rectifier K (GIRK) channels and the ATP-sensitive K (KATP) + + channels. The two-pore domain K (K2P) channels are responsible for the large background K current. Some of the actions of Ca2 + are carried out by Ca2+-sensitive K+ channels and Ca2+-sensitive Cl − channels. The latter are members of a large group of chloride channels and transporters with multiple functions. There is a large family of ATP-binding cassette (ABC) transporters some of which have a signalling role in that they extrude signalling components from the cell. One of the ABC transporters is the cystic − − fibrosis transmembrane conductance regulator (CFTR) that conducts anions (Cl and HCO3 )and contributes to the osmotic gradient for the parallel flow of water in various transporting epithelia. -

Transcriptomic Uniqueness and Commonality of the Ion Channels and Transporters in the Four Heart Chambers Sanda Iacobas1, Bogdan Amuzescu2 & Dumitru A
www.nature.com/scientificreports OPEN Transcriptomic uniqueness and commonality of the ion channels and transporters in the four heart chambers Sanda Iacobas1, Bogdan Amuzescu2 & Dumitru A. Iacobas3,4* Myocardium transcriptomes of left and right atria and ventricles from four adult male C57Bl/6j mice were profled with Agilent microarrays to identify the diferences responsible for the distinct functional roles of the four heart chambers. Female mice were not investigated owing to their transcriptome dependence on the estrous cycle phase. Out of the quantifed 16,886 unigenes, 15.76% on the left side and 16.5% on the right side exhibited diferential expression between the atrium and the ventricle, while 5.8% of genes were diferently expressed between the two atria and only 1.2% between the two ventricles. The study revealed also chamber diferences in gene expression control and coordination. We analyzed ion channels and transporters, and genes within the cardiac muscle contraction, oxidative phosphorylation, glycolysis/gluconeogenesis, calcium and adrenergic signaling pathways. Interestingly, while expression of Ank2 oscillates in phase with all 27 quantifed binding partners in the left ventricle, the percentage of in-phase oscillating partners of Ank2 is 15% and 37% in the left and right atria and 74% in the right ventricle. The analysis indicated high interventricular synchrony of the ion channels expressions and the substantially lower synchrony between the two atria and between the atrium and the ventricle from the same side. Starting with crocodilians, the heart pumps the blood through the pulmonary circulation and the systemic cir- culation by the coordinated rhythmic contractions of its upper lef and right atria (LA, RA) and lower lef and right ventricles (LV, RV). -

Spatial Distribution of Leading Pacemaker Sites in the Normal, Intact Rat Sinoa
Supplementary Material Supplementary Figure 1: Spatial distribution of leading pacemaker sites in the normal, intact rat sinoatrial 5 nodes (SAN) plotted along a normalized y-axis between the superior vena cava (SVC) and inferior vena 6 cava (IVC) and a scaled x-axis in millimeters (n = 8). Colors correspond to treatment condition (black: 7 baseline, blue: 100 µM Acetylcholine (ACh), red: 500 nM Isoproterenol (ISO)). 1 Supplementary Figure 2: Spatial distribution of leading pacemaker sites before and after surgical 3 separation of the rat SAN (n = 5). Top: Intact SAN preparations with leading pacemaker sites plotted during 4 baseline conditions. Bottom: Surgically cut SAN preparations with leading pacemaker sites plotted during 5 baseline conditions (black) and exposure to pharmacological stimulation (blue: 100 µM ACh, red: 500 nM 6 ISO). 2 a &DUGLDFIoQChDQQHOV .FQM FOXVWHU &DFQDG &DFQDK *MD &DFQJ .FQLS .FQG .FQK .FQM &DFQDF &DFQE .FQM í $WSD .FQD .FQM í .FQN &DVT 5\U .FQM &DFQJ &DFQDG ,WSU 6FQD &DFQDG .FQQ &DFQDJ &DFQDG .FQD .FQT 6FQD 3OQ 6FQD +FQ *MD ,WSU 6FQE +FQ *MG .FQN .FQQ .FQN .FQD .FQE .FQQ +FQ &DFQDD &DFQE &DOP .FQM .FQD .FQN .FQG .FQN &DOP 6FQD .FQD 6FQE 6FQD 6FQD ,WSU +FQ 6FQD 5\U 6FQD 6FQE 6FQD .FQQ .FQH 6FQD &DFQE 6FQE .FQM FOXVWHU V6$1 L6$1 5$ /$ 3 b &DUGLDFReFHSWRUV $GUDF FOXVWHU $GUDD &DY &KUQE &KUP &KJD 0\O 3GHG &KUQD $GUE $GUDG &KUQE 5JV í 9LS $GUDE 7SP í 5JV 7QQF 3GHE 0\K $GUE *QDL $QN $GUDD $QN $QN &KUP $GUDE $NDS $WSE 5DPS &KUP 0\O &KUQD 6UF &KUQH $GUE &KUQD FOXVWHU V6$1 L6$1 5$ /$ 4 c 1HXURQDOPURWHLQV -

Ion Channels
UC Davis UC Davis Previously Published Works Title THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Ion channels. Permalink https://escholarship.org/uc/item/1442g5hg Journal British journal of pharmacology, 176 Suppl 1(S1) ISSN 0007-1188 Authors Alexander, Stephen PH Mathie, Alistair Peters, John A et al. Publication Date 2019-12-01 DOI 10.1111/bph.14749 License https://creativecommons.org/licenses/by/4.0/ 4.0 Peer reviewed eScholarship.org Powered by the California Digital Library University of California S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2019/20: Ion channels. British Journal of Pharmacology (2019) 176, S142–S228 THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Ion channels Stephen PH Alexander1 , Alistair Mathie2 ,JohnAPeters3 , Emma L Veale2 , Jörg Striessnig4 , Eamonn Kelly5, Jane F Armstrong6 , Elena Faccenda6 ,SimonDHarding6 ,AdamJPawson6 , Joanna L Sharman6 , Christopher Southan6 , Jamie A Davies6 and CGTP Collaborators 1School of Life Sciences, University of Nottingham Medical School, Nottingham, NG7 2UH, UK 2Medway School of Pharmacy, The Universities of Greenwich and Kent at Medway, Anson Building, Central Avenue, Chatham Maritime, Chatham, Kent, ME4 4TB, UK 3Neuroscience Division, Medical Education Institute, Ninewells Hospital and Medical School, University of Dundee, Dundee, DD1 9SY, UK 4Pharmacology and Toxicology, Institute of Pharmacy, University of Innsbruck, A-6020 Innsbruck, Austria 5School of Physiology, Pharmacology and Neuroscience, University of Bristol, Bristol, BS8 1TD, UK 6Centre for Discovery Brain Science, University of Edinburgh, Edinburgh, EH8 9XD, UK Abstract The Concise Guide to PHARMACOLOGY 2019/20 is the fourth in this series of biennial publications. The Concise Guide provides concise overviews of the key properties of nearly 1800 human drug targets with an emphasis on selective pharmacology (where available), plus links to the open access knowledgebase source of drug targets and their ligands (www.guidetopharmacology.org), which provides more detailed views of target and ligand properties. -

Anti-KCNA5 / Kv1.5 Antibody (ARG40377)
Product datasheet [email protected] ARG40377 Package: 50 μg anti-KCNA5 / Kv1.5 antibody Store at: -20°C Summary Product Description Rabbit Polyclonal antibody recognizes KCNA5 / Kv1.5 Tested Reactivity Hu Predict Reactivity Bov Tested Application WB Host Rabbit Clonality Polyclonal Isotype IgG Target Name KCNA5 / Kv1.5 Antigen Species Human Immunogen Synthetic peptide corresponding to aa. 583-613 of Human KCNA5. (LEKCNVKAKSNVDLRRSLYALCLDTSRETDL) Conjugation Un-conjugated Alternate Names KV1.5; HK2; HPCN1; Potassium voltage-gated channel subfamily A member 5; PCN1; ATFB7; Voltage- gated potassium channel HK2; HCK1; Voltage-gated potassium channel subunit Kv1.5 Application Instructions Application table Application Dilution WB 0.1 - 0.5 µg/ml Application Note * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. Calculated Mw 67 kDa Observed Size 67 kDa Properties Form Liquid Purification Affinity purification with immunogen. Buffer 0.2% Na2HPO4, 0.9% NaCl, 0.05% Sodium azide and 5% BSA. Preservative 0.05% Sodium azide Stabilizer 5% BSA Concentration 0.5 mg/ml Storage instruction For continuous use, store undiluted antibody at 2-8°C for up to a week. For long-term storage, aliquot and store at -20°C or below. Storage in frost free freezers is not recommended. Avoid repeated freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed www.arigobio.com 1/2 before use. Note For laboratory research only, not for drug, diagnostic or other use. Bioinformation Gene Symbol KCNA5 Gene Full Name potassium channel, voltage gated shaker related subfamily A, member 5 Background Potassium channels represent the most complex class of voltage-gated ino channels from both functional and structural standpoints. -
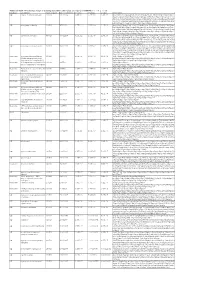
Additional Tables.Xlsx
Additional Table 6 Enriched pathways of downregulated DEGs after spinal cord injury in CRMP2KI (P < 0.05, q < 0.05) Database Description Ratio of DEGs Ratio of universe P -value P adjust q value Gene name GO neuron to neuron synapse 52/523 379/11234 1.49E-12 7.29E-10 5.98E-10 Drp2/Grik5/Tnr/Cpeb4/Cdh2/Snx27/Pde4b/Srcin1/Slitrk1/Arrb1/Add3/Cdkl5/ Neurl1a/Adgrl1/Dlg4/Pkp4/Sorbs2/Arhgap44/Pip5k1c/Ppp1r9b/Gphn/Lrrc4c/ Adam10/Ntrk2/Anks1b/Kalrn/Arhgef9/Abhd17a/Adcy1/Cnih2/Baiap2/Add1/P alm/Slc30a3/Rpl12/Dgkz/Mapk1/Map2k1/Rps18/Sptbn1/Arc/Rpl14/Atp1a1/R pl8/Nsf/Rps19/Camk2n1/Syt11/Syt1/Rps14/Nrgn/Rpl38 GO postsynaptic density 49/523 357/11234 6.77E-12 1.51E-09 1.24E-09 Drp2/Grik5/Cpeb4/Cdh2/Snx27/Pde4b/Srcin1/Slitrk1/Arrb1/Add3/Cdkl5/Neur l1a/Adgrl1/Dlg4/Pkp4/Sorbs2/Arhgap44/Pip5k1c/Ppp1r9b/Gphn/Lrrc4c/Ada m10/Ntrk2/Anks1b/Kalrn/Arhgef9/Abhd17a/Adcy1/Cnih2/Baiap2/Add1/Palm/ Rpl12/Dgkz/Mapk1/Map2k1/Rps18/Sptbn1/Arc/Rpl14/Atp1a1/Rpl8/Nsf/Rps1 9/Camk2n1/Syt11/Rps14/Nrgn/Rpl38 GO asymmetric synapse 49/523 360/11234 9.23E-12 1.51E-09 1.24E-09 Drp2/Grik5/Cpeb4/Cdh2/Snx27/Pde4b/Srcin1/Slitrk1/Arrb1/Add3/Cdkl5/Neur l1a/Adgrl1/Dlg4/Pkp4/Sorbs2/Arhgap44/Pip5k1c/Ppp1r9b/Gphn/Lrrc4c/Ada m10/Ntrk2/Anks1b/Kalrn/Arhgef9/Abhd17a/Adcy1/Cnih2/Baiap2/Add1/Palm/ Rpl12/Dgkz/Mapk1/Map2k1/Rps18/Sptbn1/Arc/Rpl14/Atp1a1/Rpl8/Nsf/Rps1 9/Camk2n1/Syt11/Rps14/Nrgn/Rpl38 GO postsynaptic specialization 50/523 382/11234 2.41E-11 2.95E-09 2.42E-09 Drp2/Grik5/Cpeb4/Cdh2/Snx27/Pde4b/Srcin1/Dlgap3/Slitrk1/Arrb1/Add3/Cdk l5/Neurl1a/Adgrl1/Dlg4/Pkp4/Sorbs2/Arhgap44/Pip5k1c/Ppp1r9b/Gphn/Lrrc4 -

Phylogenomic Analyses of KCNA Gene Clusters in Vertebrates : Why
Research article Phylogenomic analyses of KCNA gene clusters in vertebrates: why do gene clusters stay intact? Simone Hoegg and Axel Meyer* Address: Lehrstuhl für Zoologie und Evolutionsbiologie, Department of Biology, University of Konstanz, 78457 Konstanz, Germany Email: Simone Hoegg - [email protected]; Axel Meyer* - [email protected] * Corresponding author Abstract Background: Gene clusters are of interest for the understanding of genome evolution since they provide insight in large-scale duplications events as well as patterns of individual gene losses. Vertebrates tend to have multiple copies of gene clusters that typically are only single clusters or are not present at all in genomes of invertebrates. We investigated the genomic architecture and conserved non-coding sequences of vertebrate KCNA gene clusters. KCNA genes encode shaker- related voltage-gated potassium channels and are arranged in two three-gene clusters in tetrapods. Teleost fish are found to possess four clusters. The two tetrapod KNCA clusters are of approximately the same age as the Hox gene clusters that arose through duplications early in vertebrate evolution. For some genes, their conserved retention and arrangement in clusters are thought to be related to regulatory elements in the intergenic regions, which might prevent rearrangements and gene loss. Interestingly, this hypothesis does not appear to apply to the KCNA clusters, as too few conserved putative regulatory elements are retained. Results: We obtained KCNA coding sequences from basal ray-finned fishes (sturgeon, gar, bowfin) and confirmed that the duplication of these genes is specific to teleosts and therefore consistent with the fish-specific genome duplication (FSGD). -

Datasheet PB0693 Anti-KCNA5 Antibody
Product datasheet Anti-KCNA5 Antibody Catalog Number: PB0693 BOSTER BIOLOGICAL TECHNOLOGY Special NO.1, International Enterprise Center, 2nd Guanshan Road, Wuhan, China Web: www.boster.com.cn Phone: +86 27 67845390 Fax: +86 27 67845390 Email: [email protected] Basic Information Product Name Anti-KCNA5 Antibody Gene Name KCNA5 Source Rabbit IgG Species Reactivity human,mouse,rat Tested Application WB Contents 500ug/ml antibody with PBS ,0.02% NaN3 , 1mg BSA and 50% glycerol. Immunogen A synthetic peptide corresponding to a sequence at the C-terminus of human KCNA5 (583-613aa LEKCNVKAKSNVDLRRSLYALCLDTSRETDL), different from the related mouse and rat sequences by two amino acids. Purification Immunogen affinity purified. Observed MW 67KD Dilution Ratios Western blot: 1:500-2000 Storage 12 months from date of receipt,-20℃ as supplied.6 months 2 to 8℃ after reconstitution. Avoid repeated freezing and thawing Background Information Potassium voltage-gated channel, shaker-related subfamily, member 5, also known as KCNA5 or Kv1.5, is a protein that in humans is encoded by the KCNA5 gene. Potassium channels represent the most complex class of voltage-gated ion channels from both functional and structural standpoints. KCNA5 encodes a member of the potassium channel, voltage-gated, shaker-related subfamily. This member contains six membrane-spanning domains with a shaker-type repeat in the fourth segment. It belongs to the delayed rectifier class, the function of which could restore the resting membrane potential of beta cells after depolarization, thereby contributing to the regulation of insulin secretion. This gene is intronless, and the gene is clustered with genes KCNA1 and KCNA6 on chromosome 12.