N-Terminal Repeat in the Integrin A4 and A5 Subunits
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Single-Cell RNA Sequencing Reveals the Mesangial Identity and Species Diversity of Glomerular Cell Transcriptomes
ARTICLE https://doi.org/10.1038/s41467-021-22331-9 OPEN Single-cell RNA sequencing reveals the mesangial identity and species diversity of glomerular cell transcriptomes Bing He 1,11, Ping Chen 1,11, Sonia Zambrano 1,2, Dina Dabaghie1,2, Yizhou Hu 3, Katja Möller-Hackbarth1, David Unnersjö-Jess 4,5, Gül Gizem Korkut1, Emmanuelle Charrin1,2, Marie Jeansson 1, Maria Bintanel-Morcillo1,2, Anna Witasp6, Lars Wennberg7, Annika Wernerson6, Bernhard Schermer4,5, Thomas Benzing 4,5, Patrik Ernfors 3, Christer Betsholtz 1,8, Mark Lal 9, Rickard Sandberg 10 & ✉ Jaakko Patrakka 1,2 1234567890():,; Molecular characterization of the individual cell types in human kidney as well as model organisms are critical in defining organ function and understanding translational aspects of biomedical research. Previous studies have uncovered gene expression profiles of several kidney glomerular cell types, however, important cells, including mesangial (MCs) and glo- merular parietal epithelial cells (PECs), are missing or incompletely described, and a sys- tematic comparison between mouse and human kidney is lacking. To this end, we use Smart- seq2 to profile 4332 individual glomerulus-associated cells isolated from human living donor renal biopsies and mouse kidney. The analysis reveals genetic programs for all four glo- merular cell types (podocytes, glomerular endothelial cells, MCs and PECs) as well as rare glomerulus-associated macula densa cells. Importantly, we detect heterogeneity in glomerulus-associated Pdgfrb-expressing cells, including bona fide intraglomerular MCs with the functionally active phagocytic molecular machinery, as well as a unique mural cell type located in the central stalk region of the glomerulus tuft. Furthermore, we observe remarkable species differences in the individual gene expression profiles of defined glomerular cell types that highlight translational challenges in the field and provide a guide to design translational studies. -

Accumulation of Neurofascin at Nodes of Ranvier Is Regulated by a Paranodal Switch
The Journal of Neuroscience, July 22, 2020 • 40(30):5709–5723 • 5709 Cellular/Molecular Accumulation of Neurofascin at Nodes of Ranvier Is RegulatedbyaParanodalSwitch Yanqing Zhang,1* Stephanie Yuen,1 Elior Peles,3 and James L. Salzer1,2* 1Neuroscience Institute and Departments of Neuroscience and Physiology and, 2Neurology, New York University School of Medicine, New York, New York 10016, and 3Department of Molecular Cell Biology, Weizmann Institute of Science, Rehovot, 76100, Israel The paranodal junctions flank mature nodes of Ranvier and provide a barrier between ion channels at the nodes and juxta- paranodes. These junctions also promote node assembly and maintenance by mechanisms that are poorly understood. Here, we examine their role in the accumulation of NF186, a key adhesion molecule of PNS and CNS nodes. We previously showed that NF186 is initially targeted/accumulates via its ectodomain to forming PNS (hemi)nodes by diffusion trapping, whereas it is later targeted to mature nodes by a transport-dependent mechanism mediated by its cytoplasmic segment. To address the role of the paranodes in this switch, we compared accumulation of NF186 ectodomain and cytoplasmic domain constructs in WT versus paranode defective (i.e., Caspr-null) mice. Both pathways are affected in the paranodal mutants. In the PNS of Caspr-null mice, diffusion trapping mediated by the NF186 ectodomain aberrantly persists into adulthood, whereas the cyto- plasmic domain/transport-dependent targeting is impaired. In contrast, accumulation of NF186 at CNS nodes does not undergo a switch; it is predominantly targeted to both forming and mature CNS nodes via its cytoplasmic domain and requires intact paranodes. -

NFASC Rabbit Pab
Leader in Biomolecular Solutions for Life Science NFASC Rabbit pAb Catalog No.: A3053 Basic Information Background Catalog No. This gene encodes an L1 family immunoglobulin cell adhesion molecule with multiple A3053 IGcam and fibronectin domains. The protein functions in neurite outgrowth, neurite fasciculation, and organization of the axon initial segment (AIS) and nodes of Ranvier on Observed MW axons during early development. Both the AIS and nodes of Ranvier contain high 150kDa densities of voltage-gated Na+ (Nav) channels which are clustered by interactions with cytoskeletal and scaffolding proteins including this protein, gliomedin, ankyrin 3 Calculated MW (ankyrin-G), and betaIV spectrin. This protein links the AIS extracellular matrix to the 69kDa/119kDa/130-150kDa intracellular cytoskeleton. This gene undergoes extensive alternative splicing, and the full-length nature of some variants has not been determined. Category Primary antibody Applications WB Cross-Reactivity Human, Mouse, Rat Recommended Dilutions Immunogen Information WB 1:500 - 1:2000 Gene ID Swiss Prot 23114 O94856 Immunogen Recombinant fusion protein containing a sequence corresponding to amino acids 830-1100 of human NFASC (NP_001005388.2). Synonyms NFASC;NF;NRCAML Contact Product Information www.abclonal.com Source Isotype Purification Rabbit IgG Affinity purification Storage Store at -20℃. Avoid freeze / thaw cycles. Buffer: PBS with 0.02% sodium azide,50% glycerol,pH7.3. Validation Data Western blot analysis of extracts of various cell lines, using NFASC antibody (A3053) at 1:1000 dilution. Secondary antibody: HRP Goat Anti-Rabbit IgG (H+L) (AS014) at 1:10000 dilution. Lysates/proteins: 25ug per lane. Blocking buffer: 3% nonfat dry milk in TBST. -

High-Throughput Mrna Sequencing of Stromal Cells from Endometriomas and Endometrium
REPRODUCTIONRESEARCH PROOF ONLY High-throughput mRNA sequencing of stromal cells from endometriomas and endometrium Kadri Rekker1,2, Merli Saare1,2, Elo Eriste2, Tõnis Tasa3,4, Viktorija Kukuškina4,5, Anne Mari Roost2, Kristi Anderson2, Külli Samuel2, Helle Karro1,6, Andres Salumets1,2,7,8 and Maire Peters1,2 1Department of Obstetrics and Gynecology, Institute of Clinical Medicine, University of Tartu, Tartu, Estonia, 2Competence Centre on Health Technologies, Tartu, Estonia, 3Institute of Computer Science, University of Tartu, Tartu, Estonia, 4Estonian Genome Center, University of Tartu, Tartu, Estonia, 5Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia, 6Tartu University Hospital’s Women’s Clinic, Tartu, Estonia, 7Department of Biomedicine, Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu, Estonia and 8Department of Obstetrics and Gynecology, University of Helsinki and Helsinki University Hospital, Helsinki, Finland Correspondence should be addressed to K Rekker; Email: [email protected] Abstract The aetiology of endometriosis is still unclear and to find mechanisms behind the disease development, it is important to study each cell type from endometrium and ectopic lesions independently. The objective of this study was to uncover complete mRNA profiles in uncultured stromal cells from paired samples of endometriomas and eutopic endometrium. High-throughput mRNA sequencing revealed over 1300 dysregulated genes in stromal cells from ectopic lesions, including several novel genes in the context of endometriosis. Functional annotation analysis of differentially expressed genes highlighted pathways related to cell adhesion, extracellular matrix–receptor interaction and complement and coagulation cascade. Most importantly, we found a simultaneous upregulation of complement system components and inhibitors, indicating major imbalances in complement regulation in ectopic stromal cells. -

Bioinformatic Analysis Reveals the Importance of Epithelial-Mesenchymal Transition in the Development of Endometriosis
www.nature.com/scientificreports OPEN Bioinformatic analysis reveals the importance of epithelial- mesenchymal transition in the development of endometriosis Meihong Chen1,6, Yilu Zhou2,3,6, Hong Xu4, Charlotte Hill2, Rob M. Ewing2,3, Deming He1, Xiaoling Zhang1 ✉ & Yihua Wang2,3,5 ✉ Background: Endometriosis is a frequently occurring disease in women, which seriously afects their quality of life. However, its etiology and pathogenesis are still unclear. Methods: To identify key genes/ pathways involved in the pathogenesis of endometriosis, we recruited 3 raw microarray datasets (GSE11691, GSE7305, and GSE12768) from Gene Expression Omnibus database (GEO), which contain endometriosis tissues and normal endometrial tissues. We then performed in-depth bioinformatic analysis to determine diferentially expressed genes (DEGs), followed by gene ontology (GO), Hallmark pathway enrichment and protein-protein interaction (PPI) network analysis. The fndings were further validated by immunohistochemistry (IHC) staining in endometrial tissues from endometriosis or control patients. Results: We identifed 186 DEGs, of which 118 were up-regulated and 68 were down-regulated. The most enriched DEGs in GO functional analysis were mainly associated with cell adhesion, infammatory response, and extracellular exosome. We found that epithelial-mesenchymal transition (EMT) ranked frst in the Hallmark pathway enrichment. EMT may potentially be induced by infammatory cytokines such as CXCL12. IHC confrmed the down-regulation of E-cadherin (CDH1) and up-regulation of CXCL12 in endometriosis tissues. Conclusions: Utilizing bioinformatics and patient samples, we provide evidence of EMT in endometriosis. Elucidating the role of EMT will improve the understanding of the molecular mechanisms involved in the development of endometriosis. Endometriosis is a frequently occurring gynaecological disease characterised by chronic pelvic pain, dysmenor- rhea and infertility1. -

Neuronal Proteins Are Novel Components of Podocyte Major Processes and Their Expression in Glomerular Crescents Supports Their R
http://www.kidney-international.org basic research & 2012 International Society of Nephrology Neuronal proteins are novel components of podocyte major processes and their expression in glomerular crescents supports their role in crescent formation Laleh Sistani1, Patricia Q. Rodriguez1, Kjell Hultenby2, Mathias Uhlen3, Christer Betsholtz4, Hannu Jalanko5, Karl Tryggvason1, Annika Wernerson6 and Jaakko Patrakka1 1Division of Matrix Biology, Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Stockholm, Sweden; 2Division of Clinical Research Center, Department of Laboratory Medicine, Karolinska Institutet, Stockholm, Sweden; 3Department of Biotechnology, Royal Institute of Technology, Stockholm, Sweden; 4Division of Vascular Biology, Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Stockholm, Sweden; 5Hospital for Children and Adolescents, University of Helsinki, Helsinki, Finland and 6Division of Renal Medicine, Department of Clinical Science, Technology and Intervention, Karolinska Institutet, Stockholm, Sweden The podocyte has a central role in the glomerular filtration The glomerulus comprises a tangled network of capillaries barrier typified by a sophisticated morphology of highly surrounded by a urinary space and Bowman’s capsule. organized primary (major) and secondary (foot) processes. The filtration of primary urine occurs through the capillary The molecular makeup of foot processes is well wall of the glomerulus. This filtration barrier is essentially characterized, but that of major -

Neurofascin (NFASC) (NM 001160331) Human Tagged ORF Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RC228652 Neurofascin (NFASC) (NM_001160331) Human Tagged ORF Clone Product data: Product Type: Expression Plasmids Product Name: Neurofascin (NFASC) (NM_001160331) Human Tagged ORF Clone Tag: Myc-DDK Symbol: NFASC Synonyms: NEDCPMD; NF; NRCAML Vector: pCMV6-Entry (PS100001) E. coli Selection: Kanamycin (25 ug/mL) Cell Selection: Neomycin ORF Nucleotide >RC228652 representing NM_001160331 Sequence: Red=Cloning site Blue=ORF Green=Tags(s) TTTTGTAATACGACTCACTATAGGGCGGCCGGGAATTCGTCGACTGGATCCGGTACCGAGGAGATCTGCC GCCGCGATCGCC ATGGCCAGGCAGCCACCGCCGCCCTGGGTCCATGCAGCCTTCCTCCTCTGCCTCCTCAGTCTTGGCGGAG CCATCGAAATTCCTATGGATCTGACGCAGCCGCCAACCATCACCAAGCAGTCAGCGAAGGATCACATCGT GGACCCCCGTGATAACATCCTGATTGAGTGTGAAGCAAAAGGGAACCCTGCCCCCAGCTTCCACTGGACA CGAAACAGCAGATTCTTCAACATCGCCAAGGACCCCCGGGTGTCCATGAGGAGGAGGTCTGGGACCCTGG TGATTGACTTCCGCAGTGGCGGGCGGCCGGAGGAATATGAGGGGGAATATCAGTGCTTCGCCCGCAACAA ATTTGGCACGGCCCTGTCCAATAGGATCCGCCTGCAGGTGTCTAAATCTCCTCTGTGGCCCAAGGAAAAC CTAGACCCTGTCGTGGTCCAAGAGGGCGCTCCTTTGACGCTCCAGTGCAACCCCCCGCCTGGACTTCCAT CCCCGGTCATCTTCTGGATGAGCAGCTCCATGGAGCCCATCACCCAAGACAAACGTGTCTCTCAGGGCCA TAACGGAGACCTATACTTCTCCAACGTGATGCTGCAGGACATGCAGACCGACTACAGTTGTAACGCCCGC TTCCACTTCACCCACACCATCCAGCAGAAGAACCCTTTCACCCTCAAGGTCCTCACCAACCACCCTTATA ATGACTCGTCCTTAAGAAACCACCCTGACATGTACAGTGCCCGAGGAGTTGCAGAAAGAACACCAAGCTT CATGTATCCCCAGGGCACCGCGAGCAGCCAGATGGTGCTTCGTGGCATGGACCTCCTGCTGGAATGCATC -

Multiple Endocrine Neoplasia Type 1: the Potential Role of Micrornas in the Management of the Syndrome
International Journal of Molecular Sciences Review Multiple Endocrine Neoplasia Type 1: The Potential Role of microRNAs in the Management of the Syndrome Simone Donati 1, Simone Ciuffi 1 , Francesca Marini 1, Gaia Palmini 1 , Francesca Miglietta 1, Cinzia Aurilia 1 and Maria Luisa Brandi 1,2,3,* 1 Department of Experimental and Clinical Biomedical Sciences “Mario Serio”, University of Study of Florence, Viale Pieraccini 6, 50139 Florence, Italy; [email protected] (S.D.); simone.ciuffi@unifi.it (S.C.); francesca.marini@unifi.it (F.M.); gaia.palmini@unifi.it (G.P.); [email protected] (F.M.); [email protected] (C.A.) 2 Unit of Bone and Mineral Diseases, University Hospital of Florence, Largo Palagi 1, 50139 Florence, Italy 3 Fondazione Italiana Ricerca Sulle Malattie Dell’Osso (FIRMO Onlus), 50141 Florence, Italy * Correspondence: marialuisa.brandi@unifi.it; Tel.: +39-055-7946304 Received: 24 September 2020; Accepted: 12 October 2020; Published: 14 October 2020 Abstract: Multiple endocrine neoplasia type 1 (MEN1) is a rare inherited tumor syndrome, characterized by the development of multiple neuroendocrine tumors (NETs) in a single patient. Major manifestations include primary hyperparathyroidism, gastro-entero-pancreatic neuroendocrine tumors, and pituitary adenomas. In addition to these main NETs, various combinations of more than 20 endocrine and non-endocrine tumors have been described in MEN1 patients. Despite advances in diagnostic techniques and treatment options, which are generally similar to those of sporadic tumors, patients with MEN1 have a poor life expectancy, and the need for targeted therapies is strongly felt. MEN1 is caused by germline heterozygous inactivating mutations of the MEN1 gene, which encodes menin, a tumor suppressor protein. -
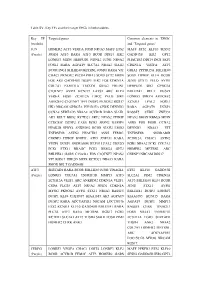
Targeted Genes Common Elements In
Table SV. Key TFs and their target DEGs in hub modules. Key TF Targeted genes Common elements in ‘DEGs’ (module) and ‘Targeted genes’ JUN HOMER2 ATF3 VEGFA FOSB NR4A3 MAFF ETS2 MAFF ETS2 KLF10 SESN2 (Purple) JOSD1 ATF3 RARA ATF3 BCOR DDIT4 IER2 GADD45B IER2 GPT2 LONRF3 MIDN HERPUD1 NDNL2 JUNB NR4A2 PHACTR3 DDIT4 ING1 SKP2 FOSL1 RARA AGPAT9 SLC7A1 NR4A2 SIAH2 CDKN1A VEGFA ATF3 BCOR ING1 BHLHE40 METRNL JOSD1 RARA VIT GRIA2 PPP1R15A BHLHE40 CHAC1 PKNOX2 PLCB4 PHF13 SOX9 SYT2 MIDN SOX9 ITPRIP KLF4 BCOR FOS AK5 GADD45B DUSP1 IER2 FOS CDKN1A JUNB STX11 PELO AVPI1 COL7A1 FAM131A TBCCD1 GRIA2 ERLIN1 HERPUD1 SIK1 GPRC5A C1QTNF7 AVPI1 KCNJ15 LATS2 ARC KLF4 BHLHE41 RELT DUSP2 VEGFA FOSB ZC3H12A LMO2 PELO SIK1 LONRF3 SPRY4 ARHGEF2 ARHGEF2 C1QTNF7 TFPI DUSP2 PKNOX2 RGS17 KCNJ15 LPAL2 FOSL1 HK1 NRCAM GPRC5A PPP1R15A CERK DENND3 RARA AGPAT9 DUSP1 CCNA2 SERTAD3 NR4A2 ACVR1B RARA SIAH1 RASSF5 CERK ZNF331 AK5 RELT BRD2 KCTD21 SKP2 NPAS2 ITPRIP NPAS2 SMOX RBM24 MIDN CCDC85C DUSP2 CARS EGR1 SESN2 RASSF5 ASNS FOS FOSB CCNA2 PIP4K2B SPRY4 ANKRD52 BCOR SIAH2 LMO2 DENND3 NR4A3 VIT TNFRSF1B ASTN2 PHACTR3 ASNS FEM1C TNFRSF1B SNORA80B CSRNP3 ITPRIP BNIP3L ATF3 ZNF331 RARA ZC3H12A CHAC1 ASTN2 VEZF1 DUSP1 SNORA80B KLF10 LPAL2 UBE2O EGR1 NR4A2 PCK1 COL7A1 PCK1 STX11 RRAGC PCK1 RBM24 GPT2 HOMER2 METRNL ARC BHLHE41 GARS C15orf41 FOS C1QTNF7 NPAS2 CSRNP3 NRCAM RGS17 VIT RGS17 UBE2O SOX9 KCTD21 NR4A3 RARA SMOX RELT GADD45B ATF3 SERTAD1 RARA BCOR BHLHE40 JUNB TINAGL1 ETS2 KLF10 GADD45B (Purple) LONRF3 COL7A1 TNFRSF1B MMP13 ATF3 SLC2A1 PIM2 CDKN1A ZC3H12A VEZF1 ARC ANKRD52 -
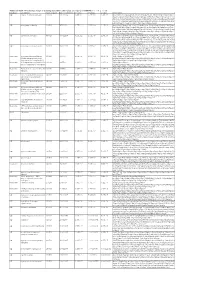
Additional Tables.Xlsx
Additional Table 6 Enriched pathways of downregulated DEGs after spinal cord injury in CRMP2KI (P < 0.05, q < 0.05) Database Description Ratio of DEGs Ratio of universe P -value P adjust q value Gene name GO neuron to neuron synapse 52/523 379/11234 1.49E-12 7.29E-10 5.98E-10 Drp2/Grik5/Tnr/Cpeb4/Cdh2/Snx27/Pde4b/Srcin1/Slitrk1/Arrb1/Add3/Cdkl5/ Neurl1a/Adgrl1/Dlg4/Pkp4/Sorbs2/Arhgap44/Pip5k1c/Ppp1r9b/Gphn/Lrrc4c/ Adam10/Ntrk2/Anks1b/Kalrn/Arhgef9/Abhd17a/Adcy1/Cnih2/Baiap2/Add1/P alm/Slc30a3/Rpl12/Dgkz/Mapk1/Map2k1/Rps18/Sptbn1/Arc/Rpl14/Atp1a1/R pl8/Nsf/Rps19/Camk2n1/Syt11/Syt1/Rps14/Nrgn/Rpl38 GO postsynaptic density 49/523 357/11234 6.77E-12 1.51E-09 1.24E-09 Drp2/Grik5/Cpeb4/Cdh2/Snx27/Pde4b/Srcin1/Slitrk1/Arrb1/Add3/Cdkl5/Neur l1a/Adgrl1/Dlg4/Pkp4/Sorbs2/Arhgap44/Pip5k1c/Ppp1r9b/Gphn/Lrrc4c/Ada m10/Ntrk2/Anks1b/Kalrn/Arhgef9/Abhd17a/Adcy1/Cnih2/Baiap2/Add1/Palm/ Rpl12/Dgkz/Mapk1/Map2k1/Rps18/Sptbn1/Arc/Rpl14/Atp1a1/Rpl8/Nsf/Rps1 9/Camk2n1/Syt11/Rps14/Nrgn/Rpl38 GO asymmetric synapse 49/523 360/11234 9.23E-12 1.51E-09 1.24E-09 Drp2/Grik5/Cpeb4/Cdh2/Snx27/Pde4b/Srcin1/Slitrk1/Arrb1/Add3/Cdkl5/Neur l1a/Adgrl1/Dlg4/Pkp4/Sorbs2/Arhgap44/Pip5k1c/Ppp1r9b/Gphn/Lrrc4c/Ada m10/Ntrk2/Anks1b/Kalrn/Arhgef9/Abhd17a/Adcy1/Cnih2/Baiap2/Add1/Palm/ Rpl12/Dgkz/Mapk1/Map2k1/Rps18/Sptbn1/Arc/Rpl14/Atp1a1/Rpl8/Nsf/Rps1 9/Camk2n1/Syt11/Rps14/Nrgn/Rpl38 GO postsynaptic specialization 50/523 382/11234 2.41E-11 2.95E-09 2.42E-09 Drp2/Grik5/Cpeb4/Cdh2/Snx27/Pde4b/Srcin1/Dlgap3/Slitrk1/Arrb1/Add3/Cdk l5/Neurl1a/Adgrl1/Dlg4/Pkp4/Sorbs2/Arhgap44/Pip5k1c/Ppp1r9b/Gphn/Lrrc4