Genetic Evidence for a Tacaribe Serocomplex Virus, Mexico
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Inventory of Mammals at Walnut Canyon, Wupatki, and Sunset Crater National Monuments
National Park Service U.S. Department of the Interior Natural Resource Program Center Inventory of Mammals at Walnut Canyon, Wupatki, and Sunset Crater National Monuments Natural Resource Technical Report NPS/SCPN/NRTR–2009/278 ON THE COVER: Top: Wupatki National Monument; bottom left: bobcat (Lynx rufus); bottom right: Wupatki pocket mouse (Perogna- thus amplus cineris) at Wupatki National Monument. Photos courtesy of U.S. Geological Survey/Charles Drost. Inventory of Mammals at Walnut Canyon, Wupatki, and Sunset Crater National Monuments Natural Resource Technical Report NPS/SCPN/NRTR—2009/278 Author Charles Drost U.S. Geological Survey Southwest Biological Science Center 2255 N. Gemini Drive Flagstaff, AZ 86001 Editing and Design Jean Palumbo National Park Service, Southern Colorado Plateau Network Northern Arizona University Flagstaff, Arizona December 2009 U.S. Department of the Interior National Park Service Natural Resource Program Center Fort Collins, Colorado The National Park Service, Natural Resource Program Center publishes a range of reports that address natural resource topics of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Technical Report Series is used to disseminate results of scientific studies in the physical, biological, and social sciences for both the advancement of science and the achievement of the National Park Service mission. The series provides contributors with a forum for displaying comprehensive data that are often deleted from journals because of page limitations. All manuscripts in the series receive the appropriate level of peer review to ensure that the information is scientifically credible, technically accurate, appropriately written for the intended audience, and designed and published in a professional manner. -
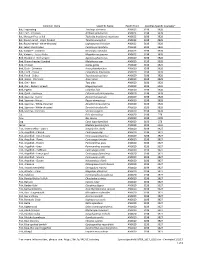
*For More Information, Please See
Common Name Scientific Name Health Point Specifies-Specific Course(s)* Bat, Frog-eating Trachops cirrhosus AN0023 3198 3928 Bat, Fruit - Jamaican Artibeus jamaicensis AN0023 3198 3928 Bat, Mexican Free-tailed Tadarida brasiliensis mexicana AN0023 3198 3928 Bat, Round-eared - stripe-headed Tonatia saurophila AN0023 3198 3928 Bat, Round-eared - white-throated Lophostoma silvicolum AN0023 3198 3928 Bat, Seba's short-tailed Carollia perspicillata AN0023 3198 3928 Bat, Vampire - Common Desmodus rotundus AN0023 3198 3928 Bat, Vampire - Lesser False Megaderma spasma AN0023 3198 3928 Bird, Blackbird - Red-winged Agelaius phoeniceus AN0020 3198 3928 Bird, Brown-headed Cowbird Molothurus ater AN0020 3198 3928 Bird, Chicken Gallus gallus AN0020 3198 3529 Bird, Duck - Domestic Anas platyrhynchos AN0020 3198 3928 Bird, Finch - House Carpodacus mexicanus AN0020 3198 3928 Bird, Finch - Zebra Taeniopygia guttata AN0020 3198 3928 Bird, Goose - Domestic Anser anser AN0020 3198 3928 Bird, Owl - Barn Tyto alba AN0020 3198 3928 Bird, Owl - Eastern Screech Megascops asio AN0020 3198 3928 Bird, Pigeon Columba livia AN0020 3198 3928 Bird, Quail - Japanese Coturnix coturnix japonica AN0020 3198 3928 Bird, Sparrow - Harris' Zonotrichia querula AN0020 3198 3928 Bird, Sparrow - House Passer domesticus AN0020 3198 3928 Bird, Sparrow - White-crowned Zonotrichia leucophrys AN0020 3198 3928 Bird, Sparrow - White-throated Zonotrichia albicollis AN0020 3198 3928 Bird, Starling - Common Sturnus vulgaris AN0020 3198 3928 Cat Felis domesticus AN0020 3198 279 Cow Bos taurus -

Blocks All Weather Bait
Brand KILLS RATS AND MICE* Blocks All Weather Bait Kills Rats, Mice, and Meadow Voles* Kills Norway Rats, Roof Rats, Cotton Rats*, Polynesian Rats*, White-throated woodrat*, Southern plains woodrat*, Mexican woodrat*, House Mice, Eastern Harvest Mice*, Golden Mice*, and Meadow Voles* Norway Rats, Roof Rats, and House Mice usually consume a lethal dose in a single night’s feeding, but it may take two or more days from time of bait consumption for them to die. KEEP OUT OF REACH OF This product is effective against anticoagulant-resistant Norway Rats, Roof Rats, and House Mice. CHILDREN Moisture Resistant • Weather Resistant CAUTION See back panel and inside leaflet for First Aid This product may only be used inside and within 100 feet of and Precautionary Statements man-made structures or inside of transport vehicles (ships, trains, or aircraft). This Product May Not Be Sold in Packaging that Holds Less Than 4 Pounds of Bait. Active Ingredient: Bromethalin (CAS No: 63333-35-7):......................0.01% Other Ingredients:............................................... 99.99% Total.................................................................100.00% EPA Reg. No. 61282-75 EPA Est. No. 61282-WI-01 Item No. 112831 BK220498-1219 *Not permitted for use against the following species in California: Cotton rat, Polynesian rat, White-throated woodrat, Southern plains woodrat, Mexican woodrat, Eastern harvest mouse, Golden mouse, Meadow vole Net Contents: 128 x 0.5 oz. (14 g) Net Weight: 4 lbs. (1.81 kg) BK 220062 (See front page of leaflet for First Aid) Bait stations are mandatory for outdoor, above-ground use. Tamper-resistant bait stations must be used wherever children, pets, non-target mammals, or birds SureKill® Brand Assault® Blocks may have access to the bait placement location. -

Presentation Abstracts/Resúmenes De Presentaciones
1 PRESENTATION ABSTRACTS/RESÚMENES DE PRESENTACIONES WILKS AWARD COMPETITION/COMPETICIÓN PARA EL PREMIO WILKS W1 BIRD OCCUPANCY IN RELATION TO HABITAT STRUCTURE IN THE OAK SAVANNA CROSS TIMBERS OF KANSAS, USA / OCUPACION DE PAJAROS EN RELACION A ESTRUCTURAS DE HABITAT EN EL ROBLE SABANA A TRAVES DE BISQUES DE KANSAS, USA Nathan S. Holoubek and William E. Jensen Department of Biological Sciences, Emporia State University, 1200 Commercial Ave, Emporia, KS 66801 Oak savanna, once widespread across central North America, has functionally vanished from most of its range. Our objective was to quantify avian habitat associations across a gradient from open-canopy oak savanna to closed-canopy forest in the Cross Timbers region of southeastern Kansas. Using 2012 point-count data, we modeled species-specific detection (p) and occupancy (ψ) probabilities against vegetative variables using program Presence. We first established the best p predictors and then used these when modeling ψ. Of 28 species modeled, ψ for 7 was strongly associated with vegetative variables. Occupancies of Contopus virens, Polioptila caerulea, and Passerina cyanea peaked at intermediate tree densities or canopy coverages. Icterus spurius, Spiza americana, and Spizella pusilla occupancies were highest at low tree densities or canopy coverages. These species might benefit from reductions in tree density within otherwise closed-canopy forest. Moderate habitat relationships with ψ were found for three other species. Point counts will resume in 2013. Quantifying bird habitat use in oak savanna will be useful in guiding future savanna restoration for avian conservation. Roble Sabana, una vez extendido a traves de Norte America central, ha funcionalmente desaparecido en la mayoria de su extension. -

Mammals of Central Mexico Juan Cruzado Cortes and Venkat Sankar (Author; [email protected]) August 5-10, 2019
Venkat Sankar 2019 1 Mammals of Central Mexico Juan Cruzado Cortes and Venkat Sankar (author; [email protected]) August 5-10, 2019 Beautiful scenery at Barrancas de Aguacatitla; Mexican Volcano Mouse; Mexican Ground Squirrel Introduction While searching for mammals in Oaxaca this March, Juan told me that a mammalogist friend of his in Tabasco, Dr. Rafael Avila Flores, had found some amazing bats in an area of karst near the state’s border with Chiapas. These included a number of impressive and distinctive species I’ve long wanted to see, like the Sword-nosed Bat and White-winged Vampire Bat. I had to visit, and with few breaks this summer thanks to academic commitments, this was the perfect choice for a long weekend’s trip. Juan suggested we spend a few days in Mexico City with another biologist friend, Melany Aguilar Lopez, to find several endemics of the Mexican Plateau, and then connect to Tabasco. And so a plan was formed! Itinerary 8/5/19: Mexico City—RB Barrancas de Metztitlan (O/N UMA Santana) 8/6/19: RB Barrancas de Metztitlan—PN el Chico (O/N Mineral de Chico) 8/7/19: PN el Chico—Tlaxco—Area Communitaria Milpa Alta (O/N San Pablo Oztotepec) 8/8/19: Milpa Alta—Villahermosa (flight)—Ejido Poana (O/N Tacotalpa) 8/9/19: Full day exploring Ejido Poana (O/N Tacotalpa) 8/10/19: Early deparature from Villahermosa Key sites RB Barrancas de Metztitlan This scenic area of deep canyons spans a diverse range of habitats from dry pine-oak forest on the rim, into high desert, and eventually tropical deciduous forest on the canyon floor. -

PROCEEDINGS of the OKLAHOMA ACADEMY of SCIENCE Volume 98 2018
PROCEEDINGS of the OKLAHOMA ACADEMY OF SCIENCE Volume 98 2018 EDITOR: Mostafa Elshahed Production Editor: Tammy Austin Business Manager: T. David Bass The Official Organ of the OKLAHOMA ACADEMY OF SCIENCE Which was established in 1909 for the purpose of stimulating scientific research; to promote fraternal relationships among those engaged in scientific work in Oklahoma; to diffuse among the citizens of the State a knowledge of the various departments of science; and to investigate and make known the material, educational, and other resources of the State. Affiliated with the American Association for the Advancement of Science. Publication Date: January 2019 ii POLICIES OF THE PROCEEDINGS The Proceedings of the Oklahoma Academy of Science contains papers on topics of interest to scientists. The goal is to publish clear communications of scientific findings and of matters of general concern for scientists in Oklahoma, and to serve as a creative outlet for other scientific contributions by scientists. ©2018 Oklahoma Academy of Science The Proceedings of the Oklahoma Academy Base and/or other appropriate repository. of Science contains reports that describe the Information necessary for retrieval of the results of original scientific investigation data from the repository will be specified in (including social science). Papers are received a reference in the paper. with the understanding that they have not been published previously or submitted for 4. Manuscripts that report research involving publication elsewhere. The papers should be human subjects or the use of materials of significant scientific quality, intelligible to a from human organs must be supported by broad scientific audience, and should represent a copy of the document authorizing the research conducted in accordance with accepted research and signed by the appropriate procedures and scientific ethics (proper subject official(s) of the institution where the work treatment and honesty). -

US EPA, Pesticide Product Label, ROZOL VOLE BAIT,12/08/2020
UNITED STATES ENVIRONMENTAL PROTECTION AGENCY WASHINGTON, DC 20460 OFFICE OF CHEMICAL SAFETY AND POLLUTION PREVENTION December 8, 2020 Kylli Paavola Sr. Compliance Specialist Liphatech, Inc. 3600 W. Elm Street Milwaukee, WI 53209 Subject: Label Amendment – Adding bushy-tailed woodrat to the label Product Name: Rozol Vole Bait EPA Registration Number: 7173-242 Application Date: June 10, 2020 Decision Number: 564027 Dear Ms. Paavola: The amended label referred to above, submitted in connection with registration under the Federal Insecticide, Fungicide and Rodenticide Act, as amended, is acceptable. This approval does not affect any conditions that were previously imposed on this registration. You continue to be subject to existing conditions on your registration and any deadlines connected with them. A stamped copy of your labeling is enclosed for your records. This labeling supersedes all previously accepted labeling. You must submit one copy of the final printed labeling before you release the product for shipment with the new labeling. In accordance with 40 CFR 152.130(c), you may distribute or sell this product under the previously approved labeling for 18 months from the date of this letter. After 18 months, you may only distribute or sell this product if it bears this new revised labeling or subsequently approved labeling. “To distribute or sell” is defined under FIFRA section 2(gg) and its implementing regulation at 40 CFR 152.3. Should you wish to add/retain a reference to the company’s website on your label, then please be aware that the website becomes labeling under the Federal Insecticide Fungicide and Rodenticide Act and is subject to review by the Agency. -

Updated and Revised Checklist of the Mammals of Oklahoma, 2019
1 Updated and Revised Checklist of the Mammals of Oklahoma, 2019 William Caire Biology Department, University of Central Oklahoma, Edmond, OK 73031 Lynda Samanie Loucks Biology Department, University of Central Oklahoma, Edmond, OK 73031 Michelle L. Haynie Biology Department, University of Central Oklahoma, Edmond, OK 73031 Brandi S. Coyner Sam Noble Oklahoma Museum of Natural History, Department of Mammalogy, Norman, OK 73072 Janet K. Braun Sam Noble Oklahoma Museum of Natural History, Department of Mammalogy, Norman, OK 73072 Abstract: An updated list of the mammals of Oklahoma was compiled from literature records, sight records, and museum specimens. A total of 108 native species, 4 extirpated species, and 5 introduced/exotic species are reported. jugossicularis, and Perognathus merriami), not Introduction included in the most recent checklist of Choate and Jones (1998), have been verified as occurring in the state. Choate and Jones (1998) included In a checklist of mammals of Oklahoma the domestic dog and cat as introduced/exotic (Caire et al. 1989), a total of 106 species of species which we did not. This document has mammals were listed as occurring in Oklahoma, been created in part to assist those working with including 4 extirpated and 4 introduced species. the many different and varied aspects related to In 1998, an updated checklist was published the state’s mammals. It will provide a common (Choate and Jones 1998) listing 111 species point of reference and terminology. of mammals including 4 extirpated and 7 introduced/exotic species. Since the publication Methods by Caire et al. (1989) and the updated checklist of Choate and Jones (1998), there have been To compile the updated list, we began with several changes in distributional occurrences Caire et al. -

US EPA, Pesticide Product Label, Bromethalin Soft Bait II,08/02/2021
U.S. ENVIRONMENTAL PROTECTION AGENCY EPA Reg. Number: Date of Issuance: Office of Pesticide Programs Registration Division (7505P) 7173-310 8/2/21 1200 Pennsylvania Ave., N.W. Washington, D.C. 20460 NOTICE OF PESTICIDE: Term of Issuance: X Registration Reregistration Conditional (under FIFRA, as amended) Name of Pesticide Product: Bromethalin Soft Bait II Name and Address of Registrant (include ZIP Code): Kylli Paavoli Liphatech, Inc. 3600 W. Elm Street Milwaukee, WI 53209 Note: Changes in labeling differing in substance from that accepted in connection with this registration must be submitted to and accepted by the Registration Division prior to use of the label in commerce. In any correspondence on this product always refer to the above EPA registration number. On the basis of information furnished by the registrant, the above named pesticide is hereby registered under the Federal Insecticide, Fungicide and Rodenticide Act. Registration is in no way to be construed as an endorsement or recommendation of this product by the Agency. In order to protect health and the environment, the Administrator, on his motion, may at any time suspend or cancel the registration of a pesticide in accordance with the Act. The acceptance of any name in connection with the registration of a product under this Act is not to be construed as giving the registrant a right to exclusive use of the name or to its use if it has been covered by others. This product is conditionally registered in accordance with FIFRA section 3(c)(7)(A). You must comply with the following conditions: 1. Submit and/or cite all data required for registration/reregistration/registration review of your product under FIFRA when the Agency requires all registrants of similar products to submit such data. -

Mammals of Colorado, Second Edition
1 Environments of Colorado Mammals are a familiar and important component of understand the distribution and abundance of mammals Earth’s biodiversity. Biodiversity is the kinds of organisms and the details of their daily lives we must fi rst understand and their genetic and ecological relationships—an evolu- the resource base, the mosaic of Colorado’s environments tionary and ecological phenomenon in space and time (E. in space and time. Wilson 1992). The mammalian fauna of Colorado is a fas- cinating piece of that whole. To understand the diversity of mammals we need to have a perspective of the ecosphere more generally. Such a perspective is the purpose of this Geography chapter, with a focus on environments of Colorado. Colorado is known for its scenic beauty—from majes- From the standpoint of political geography, Colorado is tic mountain peaks and rushing white rivers tumbling simple: it is roughly rectangular (if we neglect some minor down dark canyons, to red-rock deserts and ceaselessly old surveyors’ errors and the fact that Earth is spherical), shifting sand dunes, to the expansive sweep of the short- measuring approximately 607 km by 444 km (377 by 276 grass prairie. Grandeur is wherever we stop to appreciate mi.) and encompassing some 270,000 km2 (104,000 sq. mi.). it, at every scale, from canyons carved in crystalline rocks Colorado lies between approximately 102° and 109° west 2 billion years old, to bold peaks sculpted by the glaciers longitude and 37° and 41° north latitude, and is subdi- of the last Ice Age, to last night’s furtive trail of a mouse vided into 64 counties (Map 1-1). -

Stable Grassland Ecosystem
This file was created by scanning the printed publication. Errors identified by the software have been corrected; however, some errors may remain. Chapter 5 Desert Grassland and Shrubland Ecosystems Samuel R. Loftin, USDA Forest Service, Rocky Mountain Forest and Range Experiment Station, Albuquerque, New Mexico Richard Aguilar, Sandia National Laboratories, Albuquerque, New Mexico Alice L. Chung-MacCoubrey, USDA Forest Service, . Rocky Mountain Forest and Range Experiment Station, Albuquerque, New MexIco Wayne A. Robbie, USDA Forest Service, Watershed an~ Air Management Staff, Region 3, Albuquerque, New MexIco INTRODUCTION taken from a review by Dick-Peddie (1993). The ex tensive grasslands and shrublands of North America The productivity, stability, and health of the Middle developed in response to climate change and the Rio Grande Basin, arid and semiarid grassland and uplifting of the western mountain ranges, includ.ing shrub land ecosystems depend upon complex inter the Rocky, Cascade, and Sierra Nevada Mountains, actions. These relationships occur between factors which began in the early to mid-Tertiary period (65- such as climate, domestic livestock, and wildlife use, 26 million years ago). The vegetation of North and human activities such as urban development, ag America early in the Tertiary period has been riculture, and recreation. These grassland/ shrubland grouped into three major geoflora.s. The Artc~ ecosystems are particularly sensitive to change because Tertiary Geoflora occupied cool, mOIst, upper latI they depend highly upon water availability. tudes, the Neotropical-Tertiary Geoflora occupied the Southwestern rangelands experienced heavy live warm, moist,lower latitudes, and the Madro-Tertiary stock grazing and human activities over the past cen Geoflora occupied intermediate, drier regions. -

Hubbell Trading Post National Historic Site
Final Report for 2003 and 2004 Mammal Inventories on Selected National Park Service Southern Colorado Plateau Network Parks: Hubbell Trading Post National Historic Site January 2005 Prepared by: Shauna Haymond, Holistic Wildlife Services NM, LLC, 112 Hampton Roads Avenue, Hampton, Virginia 23661, and Richard E. Sherwin, Department of Biology, Chemistry and Environmental Science, Christopher Newport University, 1 University Place, Newport News, Virginia 23606-2998 Submitted to: Navajo Nation Department of Fish and Wildlife, P. O. Box 1480, Window Rock, AZ 96515 ABSTRACT Holistic Wildlife Services NM was contracted by the Navajo Nation Department of Fish and Wildlife to conduct biological inventories for mammals at Hubbell Trading Post National Historic Site (HUTR) as part of the National Park Service Inventory and Monitoring Program. The goals of this study were to document at least 90% of the mammals using verifiable documentation and taxa-specific field surveys, provide distributional information, estimates of species richness, and relative abundance of mammals, and provide baseline information and make recommendations to develop future management and monitoring schemes of zoological resources. There had been no baseline mammal work conducted at HUTR prior to these surveys. A total of 23 mammal species were estimated to inhabit the park based on species-area models; however we estimated 39 species for HUTR based on known specific ranges and available museum records. Field inventories extended from 26 June to 28 August 2003, and 10 May to 17 June 2004. We used a variety of survey methods including live-trapping, mist netting and acoustic surveys for bats, track-scat surveys, and opportunistic observations.