Effects of Taxon Sampling on Molecular Dating for Within-Genus Divergence Events, When Deep Fossils Are Used for Calibration Richard I
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Macadamia Integrifolia HAES 741)
bioRxiv preprint doi: https://doi.org/10.1101/2020.05.25.114009; this version posted May 27, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Chromosome-scale assembly and annotation of the macadamia genome (Macadamia integrifolia HAES 741) Catherine J. Nock1†*, Abdul Baten1,2†, Ramil Mauleon1, Kirsty S. Langdon1, Bruce Topp3, Craig Hardner3, Agnelo Furtado3, Robert J. Henry3 and Graham J. King1 1Southern Cross Plant Science, Southern Cross University, Lismore NSW 2480, Australia 2AgResearch NZ, Grasslands Research Centre, Palmerston North 4442, New Zealand 3Queensland Alliance for Agriculture and Food Innovation, University of Queensland, St Lucia QLD 4069, Australia †These authors contributed equally to this work *Corresponding author, [email protected] ORCID 0000-0001-5609-4681 Abstract Macadamia integrifolia is a representative of the large basal eudicot family Proteaceae and the main progenitor species of the Australian native nut crop macadamia. Since its commercialisation in Hawaii fewer than 100 years ago, global production has expanded rapidly. However, genomic resources are limited in comparison to other horticultural crops. The first draft assembly of M. integrifolia had good coverage of the functional gene space but its high fragmentation has restricted its use in comparative genomics and association studies. Here we have generated an improved assembly of cultivar HAES 741 (4,094 scaffolds, 745 Mb, N50 413 kb) using a combination of Illumina paired and PacBio long read sequences. -

Plant List 2016
Established 1990 PLANT LIST 2016 European mail order website www.crug-farm.co.uk CRÛG FARM PLANTS • 2016 Welcome to our 2016 list hope we can tempt you with plenty of our old favourites as well as some exciting new plants that we have searched out on our travels. There has been little chance of us standing still with what has been going on here in 2015. The year started well with the birth of our sixth grandchild. January into February had Sue and I in Colombia for our first winter/early spring expedition. It was exhilarating, we were able to travel much further afield than we had previously, as the mountainous areas become safer to travel. We are looking forward to working ever closer with the Colombian institutes, such as the Medellin Botanic Gardens whom we met up with. Consequently we were absent from the RHS February Show at Vincent Square. We are finding it increasingly expensive participating in the London shows, while re-branding the RHS February Show as a potato event hardly encourages our type of customer base to visit. A long standing speaking engagement and a last minute change of date, meant that we missed going to Fota near Cork last spring, no such problem this coming year. We were pleasantly surprised at the level of interest at the Trgrehan Garden Rare Plant Fair, in Cornwall. Hopefully this will become an annual event for us, as well as the Cornwall Garden Society show in April. Poor Sue went through the wars having to have a rush hysterectomy in June, after some timely results revealed future risks. -

Persoonia Mollis Subsp. Revolute
NSW Threatened Species Scientific Committee Conservation Assessment of Persoonia mollis subsp. revoluta S.Krauss & L.A.S.Johnson (Proteaceae) C Simpson, May 2019 NSW Threatened Species Scientific Committee Persoonia mollis R.Br. subsp. revoluta S.Krauss & L.A.S.Johnson (Proteaceae) Distribution: Endemic to NSW Current EPBC Act Status: Not listed Current NSW BCC Act Status: Not Listed Conservation Advice: Persoonia mollis subsp. revoluta Summary of Conservation Assessment Persoonia mollis subsp. revoluta is found to be eligible for listing as Vulnerable under IUCN Criteria B1ab(i,ii,iii,iv,v)+2ab(i,ii,iii,iv,v) (IUCN 2017). The main reasons for this taxon being eligible are i) highly restricted geographic range – EOO = 1100 km2, AOO = 104 km2; ii) it is known from only seven locations; and iii) a continuing decline is inferred in the extent of occurrence, area of occupancy, area of habitat, number of locations and number of mature individuals, as a result of low frequency of fire preventing natural regeneration. Description and Taxonomy Persoonia mollis subsp. revoluta S.Krauss & L.A.S.Johnson (family Proteaceae) is a prostrate to decumbent shrub, 10-50 cm high, up to 4 m diameter; leaves glossy-green, pliable but not soft, almost fleshy, elliptical to oblong-ovate to oblong-Ianceolate, obtuse (to rarely acute), 2.5–4 cm long, 4–10 (–15) mm wide, sparsely silky-pubescent to glabrous on the undersurface when young, the longest hairs c. 0.7 mm long, the midvein obscure or (rarely) prominent, the margins revolute; buds sparsely silky pubescent to ± glabrous, the hairs 0.3–1 mm long, pale (Krauss and Johnson 1991; PlantNET 2019). -

A Compilation and Analysis of Food Plants Utilization of Sri Lankan Butterfly Larvae (Papilionoidea)
MAJOR ARTICLE TAPROBANICA, ISSN 1800–427X. August, 2014. Vol. 06, No. 02: pp. 110–131, pls. 12, 13. © Research Center for Climate Change, University of Indonesia, Depok, Indonesia & Taprobanica Private Limited, Homagama, Sri Lanka http://www.sljol.info/index.php/tapro A COMPILATION AND ANALYSIS OF FOOD PLANTS UTILIZATION OF SRI LANKAN BUTTERFLY LARVAE (PAPILIONOIDEA) Section Editors: Jeffrey Miller & James L. Reveal Submitted: 08 Dec. 2013, Accepted: 15 Mar. 2014 H. D. Jayasinghe1,2, S. S. Rajapaksha1, C. de Alwis1 1Butterfly Conservation Society of Sri Lanka, 762/A, Yatihena, Malwana, Sri Lanka 2 E-mail: [email protected] Abstract Larval food plants (LFPs) of Sri Lankan butterflies are poorly documented in the historical literature and there is a great need to identify LFPs in conservation perspectives. Therefore, the current study was designed and carried out during the past decade. A list of LFPs for 207 butterfly species (Super family Papilionoidea) of Sri Lanka is presented based on local studies and includes 785 plant-butterfly combinations and 480 plant species. Many of these combinations are reported for the first time in Sri Lanka. The impact of introducing new plants on the dynamics of abundance and distribution of butterflies, the possibility of butterflies being pests on crops, and observations of LFPs of rare butterfly species, are discussed. This information is crucial for the conservation management of the butterfly fauna in Sri Lanka. Key words: conservation, crops, larval food plants (LFPs), pests, plant-butterfly combination. Introduction Butterflies go through complete metamorphosis 1949). As all herbivorous insects show some and have two stages of food consumtion. -

IV. on the Proteaceć of Jussieu. by Mr. Robert Brown, Lib. LS
IV. On the Proteacea of Jussieu. By -Mr. Robert Brown, Lib. L.S. Read Jan. 17, 1809. THELinnean system of botany, though confessedly artificial, has not only contributed more than all others to facilitate tlie knowledge of species, but, by constantly directing the attention to those essential parts of the flower on which it is founded, has made us acquainted with more of their important modific-a t’ ions than we probably should have known, had it not been generally adopted, and has thus laid a more solid foundation for the esta- blishment of a natural arrangement, the superior importance of which no one has been inore fully impressed with than Linnzus hiinself. There are still, however, certain circumstances respccting the stamina and pistilla, which appear to iiie to havc been much less attended to than they deserve, both by Linneus and succeeding botanists. What I chiefly allude to is the state of these organs before the expansion of the flower. Tlie utility of ascertaining the internal condition of the ovarium before fecundation will liardly be called in question, now that the immortal worlis of Gxrtner and Jussieu hare demonstrated the necessity of minutely studying the fruits of plants in attempting to arrange tlicin ac- cording to tlic sum of their affinities, as in many cases the true nature of tlie ripc fruit, cspecially witli respect to the placenta- tion of the seeds, can oiily be detcrniined by this mc;~ns. Its importance is indeed expressly inculcated by many l~ot:inists, Tf’llO, 16 Mr. BROWN,on the Proteacee of Jussieu. -

Bio 308-Course Guide
COURSE GUIDE BIO 308 BIOGEOGRAPHY Course Team Dr. Kelechi L. Njoku (Course Developer/Writer) Professor A. Adebanjo (Programme Leader)- NOUN Abiodun E. Adams (Course Coordinator)-NOUN NATIONAL OPEN UNIVERSITY OF NIGERIA BIO 308 COURSE GUIDE National Open University of Nigeria Headquarters 14/16 Ahmadu Bello Way Victoria Island Lagos Abuja Office No. 5 Dar es Salaam Street Off Aminu Kano Crescent Wuse II, Abuja e-mail: [email protected] URL: www.nou.edu.ng Published by National Open University of Nigeria Printed 2013 ISBN: 978-058-434-X All Rights Reserved Printed by: ii BIO 308 COURSE GUIDE CONTENTS PAGE Introduction ……………………………………......................... iv What you will Learn from this Course …………………............ iv Course Aims ……………………………………………............ iv Course Objectives …………………………………………....... iv Working through this Course …………………………….......... v Course Materials ………………………………………….......... v Study Units ………………………………………………......... v Textbooks and References ………………………………........... vi Assessment ……………………………………………….......... vi End of Course Examination and Grading..................................... vi Course Marking Scheme................................................................ vii Presentation Schedule.................................................................... vii Tutor-Marked Assignment ……………………………….......... vii Tutors and Tutorials....................................................................... viii iii BIO 308 COURSE GUIDE INTRODUCTION BIO 308: Biogeography is a one-semester, 2 credit- hour course in Biology. It is a 300 level, second semester undergraduate course offered to students admitted in the School of Science and Technology, School of Education who are offering Biology or related programmes. The course guide tells you briefly what the course is all about, what course materials you will be using and how you can work your way through these materials. It gives you some guidance on your Tutor- Marked Assignments. There are Self-Assessment Exercises within the body of a unit and/or at the end of each unit. -

NEWSLETTER 140, January 2018
No. 140 Irish Garden Plant Society Newsletter January 2018 Irish Heritage Daffodils Irish Heritage Daffodils IGPS Newsletter January 2018 Irish Heritage Daffodils Editorial Irish Heritage Daffodils Irish Heritage Daffodils Narcissus ‘Border Beauty’ Mary Montaut, Leinster Branch IGPS Narcissus ‘Border Beauty’ The chilly, bright winter weather just after Christmas made me really appreciate some scented subjects in the garden, especially my favourite Daphne bholua ‘Jacqueline Postil’. I was extremely fortunate many years ago to attend a propagation workshop with IGPS at Kinsealy, and they had rooted cuttings of this glorious plant. I bought one and I have adored it ever since. However, I have recently fallen in love just as passionately with another winter-scented shrub and this one, I believe, might be adopted by IGPS as one of our ‘Irish Heritage’ plants, because it is named after an Irish botanist. The shrub is Edgeworthia chrysantha - the golden-headed Edgeworthia. It belongs to the same family as the Daphne, Thymelaeaceae, and originates from the China - Nepal border area. It is naturalized in Japan, where it was planted in the late sixteenth century for paper making and is called the Paper Bush (Mitsumata). There is also an orange- flowered variety called Akebono which is said to be a smaller shrub, but I have never be lucky enough to see this one. It was first classified in 1841, and named in honour Michael Pakenham Edgeworth. He was a younger brother of the novelist, Maria Edgeworth (of Castle Rackrent fame) and lived and worked in India most of his life. However, I feel we should salute his work, and recommend this superb and tolerant shrub. -

Boxwood Blight
PLANT AND PEST BP-203-W DIAGNOSTIC LABORATORY ppdl.purdue.edu Boxwood Blight Gail Ruhl Purdue Botany and Plant Pathology - ag.purdue.edu/BTNY Tom Creswell Janna Beckerman Introduction Boxwood blight is a fungal disease caused by Calonectria pseudonaviculata (previously called Cylindrocladium pseudonaviculatum or Cylindrocladium buxicola). This fungus is easily transported in the nursery industry and can be moved on infected plants that do not show any symptoms at the time of shipment as well as on shoots of infected boxwood greenery tucked into evergreen Christmas wreaths. Boxwood blight has become a serious threat to nursery production and to boxwoods in the landscape, which has prompted several states to take regulatory action. This publication provides information about boxwood blight and management options. Disease Distribution Boxwood blight was first reported in the United Kingdom in the mid 1990s. It is now widespread throughout most of Europe and was also discovered in New Zealand in 1998. Boxwood blight was confirmed for the first time in North America in October 2011 on samples collected in North Carolina and Connecticut. Since this first U.S. detection, boxwood blight has been reported in more than 20 states and three Canadian provinces. Symptoms and Signs The fungus that causes boxwood blight can infect all aboveground portions of the shrub. Symptoms begin as dark leaf spots that coalesce to form brown blotches (Figure 1). The undersides of infected leaves will show white sporulation of the boxwood blight Boxwood Blight BP-203-W fungus following periods of high humidity (Figure 2). Boxwood blight causes rapid defoliation, which usually starts on the lower branches and moves upward in the canopy (Figure 3). -

Reconstructing the Basal Angiosperm Phylogeny: Evaluating Information Content of Mitochondrial Genes
55 (4) • November 2006: 837–856 Qiu & al. • Basal angiosperm phylogeny Reconstructing the basal angiosperm phylogeny: evaluating information content of mitochondrial genes Yin-Long Qiu1, Libo Li, Tory A. Hendry, Ruiqi Li, David W. Taylor, Michael J. Issa, Alexander J. Ronen, Mona L. Vekaria & Adam M. White 1Department of Ecology & Evolutionary Biology, The University Herbarium, University of Michigan, Ann Arbor, Michigan 48109-1048, U.S.A. [email protected] (author for correspondence). Three mitochondrial (atp1, matR, nad5), four chloroplast (atpB, matK, rbcL, rpoC2), and one nuclear (18S) genes from 162 seed plants, representing all major lineages of gymnosperms and angiosperms, were analyzed together in a supermatrix or in various partitions using likelihood and parsimony methods. The results show that Amborella + Nymphaeales together constitute the first diverging lineage of angiosperms, and that the topology of Amborella alone being sister to all other angiosperms likely represents a local long branch attrac- tion artifact. The monophyly of magnoliids, as well as sister relationships between Magnoliales and Laurales, and between Canellales and Piperales, are all strongly supported. The sister relationship to eudicots of Ceratophyllum is not strongly supported by this study; instead a placement of the genus with Chloranthaceae receives moderate support in the mitochondrial gene analyses. Relationships among magnoliids, monocots, and eudicots remain unresolved. Direct comparisons of analytic results from several data partitions with or without RNA editing sites show that in multigene analyses, RNA editing has no effect on well supported rela- tionships, but minor effect on weakly supported ones. Finally, comparisons of results from separate analyses of mitochondrial and chloroplast genes demonstrate that mitochondrial genes, with overall slower rates of sub- stitution than chloroplast genes, are informative phylogenetic markers, and are particularly suitable for resolv- ing deep relationships. -

Brisbane Native Plants by Suburb
INDEX - BRISBANE SUBURBS SPECIES LIST Acacia Ridge. ...........15 Chelmer ...................14 Hamilton. .................10 Mayne. .................25 Pullenvale............... 22 Toowong ....................46 Albion .......................25 Chermside West .11 Hawthorne................. 7 McDowall. ..............6 Torwood .....................47 Alderley ....................45 Clayfield ..................14 Heathwood.... 34. Meeandah.............. 2 Queensport ............32 Trinder Park ...............32 Algester.................... 15 Coopers Plains........32 Hemmant. .................32 Merthyr .................7 Annerley ...................32 Coorparoo ................3 Hendra. .................10 Middle Park .........19 Rainworth. ..............47 Underwood. ................41 Anstead ....................17 Corinda. ..................14 Herston ....................5 Milton ...................46 Ransome. ................32 Upper Brookfield .......23 Archerfield ...............32 Highgate Hill. ........43 Mitchelton ...........45 Red Hill.................... 43 Upper Mt gravatt. .......15 Ascot. .......................36 Darra .......................33 Hill End ..................45 Moggill. .................20 Richlands ................34 Ashgrove. ................26 Deagon ....................2 Holland Park........... 3 Moorooka. ............32 River Hills................ 19 Virginia ........................31 Aspley ......................31 Doboy ......................2 Morningside. .........3 Robertson ................42 Auchenflower -
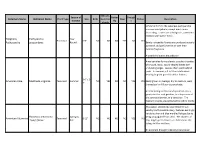
Common Name Botanical Name Alleghany
Attracts Season of Butter Drough Common Name Botanical Name Plant Type Size Birds Hummin Deer Native Description Interest fly t g birds Similar in form to the Japanese pachysandra one sees everywhere, except much more interesting. Leaves are a dull green, sometimes mottled with lighter flecks. Alleghany Pachysandra Year Perennial 6-8" NO NO NO YES NO YES Pachysandra procumbens Round Barely noticeable flowers are produced as early as March and perfume the air with their delicate fragrance. A wonderful native groundcover. American aloe forms a lovely succulent rosette of smooth, waxy, sword-shaped leaves with undulating edges. Leaves often sport reddish spots. In summer, a 3 to 5 foot stalk arises bearing fragrant greenish-white flowers. 3-6' x 2- American Aloe Manfreda virginica Perennial Summer NO YES NO NO YES YES Easily grown in average, dry to medium, well- 3' drained soil in full sun to part shade. An interesting architectural specimen, it is a good plant for rock gardens, in a dry corner of the perennial border, or a container. The fragrant blooms are pollinated by sphinx moths. This native, selected by Dale Hendrick's at nearby North Creek Nursery, features excitingly variable silver and blue marbled foliage due to Heuchera americana Spring to being propagated from seed. The clusters of American Alumroot Perennial 8-12" NO NO NO NO YES NO 'Dale's Strain' Fall tiny, bright green flowers are held above the foliage in May and June. An excellent drought tolerant groundcover. Viburnum trilobum is a native deciduous shrub to the northeastern and northwestern United States. -

Dispersal Effects on Species Distribution and Diversity Across Multiple Scales in the Southern Appalachian Mixed Mesophytic Flora
DISPERSAL EFFECTS ON SPECIES DISTRIBUTION AND DIVERSITY ACROSS MULTIPLE SCALES IN THE SOUTHERN APPALACHIAN MIXED MESOPHYTIC FLORA Samantha M. Tessel A dissertation submitted to the faculty at the University of North Carolina at Chapel Hill in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Ecology in the Curriculum for the Environment and Ecology. Chapel Hill 2017 Approved by: Peter S. White Robert K. Peet Alan S. Weakley Allen H. Hurlbert Dean L. Urban ©2017 Samantha M. Tessel ALL RIGHTS RESERVED ii ABSTRACT Samantha M. Tessel: Dispersal effects on species distribution and diversity across multiple scales in the southern Appalachian mixed mesophytic flora (Under the direction of Peter S. White) Seed and spore dispersal play important roles in the spatial distribution of plant species and communities. Though dispersal processes are often thought to be more important at larger spatial scales, the distribution patterns of species and plant communities even at small scales can be determined, at least in part, by dispersal. I studied the influence of dispersal in southern Appalachian mixed mesophytic forests by categorizing species by dispersal morphology and by using spatial pattern and habitat connectivity as predictors of species distribution and community composition. All vascular plant species were recorded at three nested sample scales (10000, 1000, and 100 m2), on plots with varying levels of habitat connectivity across the Great Smoky Mountains National Park. Models predicting species distributions generally had higher predictive power when incorporating spatial pattern and connectivity, particularly at small scales. Despite wide variation in performance, models of locally dispersing species (species without adaptations to dispersal by wind or vertebrates) were most frequently improved by the addition of spatial predictors.