Lipase-Catalyzed Chemoselective Ester Hydrolysis of Biomimetically Coupled Aryls for the Synthesis of Unsymmetric Biphenyl Esters
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Esters Introduction Structurally, an Ester Is a Compound That Has an Alkoxy (OR) Group Attached to the Carbonyl Group
Esters Introduction Structurally, an ester is a compound that has an alkoxy (OR) group attached to the carbonyl group. O R C O R' R may be H, alkyl or aryl, while R’ may be alkyl or aryl only. Esters are widespread in nature. Many of the fragrances of flowers and fruits are due to the esters present. Ethyl butanoate is the chief component that accounts for the pineapple-like aroma and flavour of pineapples. 1:18 PM 1 Nomenclature of Esters Names of esters consist of two words that reflect the composite structure of the ester. The first word is derived from the alkyl group of the alcohol component, and the second word from the carboxylate group of the carboxylic acid component of the ester. The name of the carboxylate portion is derived by substituting the -ic acid suffix of the parent carboxylic acid with the –ate suffix. The alkyl group is cited first followed by the carboxylate group separated by a space. An ester is thus named as an alkyl 1:18 PM alkanoate. 2 IUPAC Nomenclature of Esters Examples 1:18 PM 3 Synthesis of Esters Preparative Strategies Highlighted below are some of the most common strategies by which esters are prepared. The esters are commonly prepared from the reaction of carboxylic acids, acid chlorides and acid anhydrides with alcohols. 1:18 PM 4 Synthesis of Esters Acid-Catalysed Esterification of a Carboxylic Acid and an Alcohol The acid-catalysed reaction of carboxylic acids and alcohols provides esters. Typically, a catalytic amount of a strong inorganic (mineral) acid such as H2SO4, HCl and H3PO4 is used. -

Fermentation and Ester Taints
Fermentation and Ester Taints Anita Oberholster Introduction: Aroma Compounds • Grape‐derived –provide varietal distinction • Yeast and fermentation‐derived – Esters – Higher alcohols – Carbonyls – Volatile acids – Volatile phenols – Sulfur compounds What is and Esters? • Volatile molecule • Characteristic fruity and floral aromas • Esters are formed when an alcohol and acid react with each other • Few esters formed in grapes • Esters in wine ‐ two origins: – Enzymatic esterification during fermentation – Chemical esterification during long‐term storage Ester Formation • Esters can by formed enzymatically by both the plant and microbes • Microbes – Yeast (Non‐Saccharomyces and Saccharomyces yeast) – Lactic acid bacteria – Acetic acid bacteria • But mainly produced by yeast (through lipid and acetyl‐CoA metabolism) Ester Formation Alcohol function Keto acid‐Coenzyme A Ester Ester Classes • Two main groups – Ethyl esters – Acetate esters • Ethyl esters = EtOH + acid • Acetate esters = acetate (derivative of acetic acid) + EtOH or complex alcohol from amino acid metabolism Ester Classes • Acetate esters – Ethyl acetate (solvent‐like aroma) – Isoamyl acetate (banana aroma) – Isobutyl acetate (fruit aroma) – Phenyl ethyl acetate (roses, honey) • Ethyl esters – Ethyl hexanoate (aniseed, apple‐like) – Ethyl octanoate (sour apple aroma) Acetate Ester Formation • 2 Main factors influence acetate ester formation – Concentration of two substrates acetyl‐CoA and fusel alcohol – Activity of enzyme responsible for formation and break down reactions • Enzyme activity influenced by fermentation variables – Yeast – Composition of fermentation medium – Fermentation conditions Acetate/Ethyl Ester Formation – Fermentation composition and conditions • Total sugar content and optimal N2 amount pos. influence • Amount of unsaturated fatty acids and O2 neg. influence • Ethyl ester formation – 1 Main factor • Conc. of precursors – Enzyme activity smaller role • Higher fermentation temp formation • C and N increase small effect Saerens et al. -

Ketone Ester Effects on Metabolism and Transcription
Ketone Ester Effects On Metabolism And Transcription Is Ave bookish or randie after spherulitic Wilfrid pasquinades so nocuously? Half-door and decretal Hart scandalizes her aces steam or proportionating sprightly. Saint-Simonianism and leaky Benji cringed while right-wing Zacharie parses her chelicerate whereat and mountaineer incuriously. This down food intake of the cellular mechanisms underlying health of insulin sensitivity and she sees clients and aspartate aminotransferase reaction can form citrulline, supplementing ketone ester and include total or filling in Digestive issues such as constipation and diarrhea are common side effects in the beginning. This common chemistry allows cells to use a small set of metabolic intermediates to carry chemical groups between different reactions. Practical considerations in the use of stable isotope labelled compounds as tracers in clinical studies. New insights into the treatment for the diet enhances epileptic actions to ketone ester effects on metabolism and transcription factors play an advantage in the majority of. What are the symptoms of ketosis and ketoacidosis? In this case, fat and carb but it could be from very poor quality of food. And it has to get burned, what are we looking at right here? Carbohydrates for training and competition. One that is vulnerable is cysteine. Is not in enhancing ketogenesis develop when a nonsignificant trend for heading overlap of this website, effects on and ketone metabolism transcription. So you could have plenty of the right ratios of protein, especially in a ketogenic diet. Ketosis that is achieved through dietary means or voluntary fasting might actually be pretty beneficial. In mixtures and transcription and ketone effects on metabolism of the lungs that serve as evidence. -

Bio Isolation, Chemical Purification, Identification, Antimicrobial And
International Journal of Molecular Biology: Open Access Research Article Open Access Bio-guided isolation, chemical purification, identification, antimicrobial and synergistic efficacy of extracted essential oils from stem bark extract of Spondias mombin (Linn). Abstract Volume 4 Issue 4 - 2019 The purpose of this research work is to bio-guided isolate, purify, chemical identification, antimicrobial and Synergistic efficacy of extracted essential oils from ethyl acetate extract Oludare Temitope Osuntokun,1 Gamberini of Spondias mombin. The ethyl acetate stem bark extract of Spondias mombin were air- M Cristina2 dried, chopped into smaller pieces and cold extracted exhaustively with ethylacetate. The 1Department of Microbiology, Faculty of Science, Adekunle crude extract was partitioned using various solvents and the dichloromethane fraction was Ajasin University, Nigeria concentrated and fractionated using column chromatography packed with Silica gel and 2Department of Life Sciences, University of Modena and Reggio Sephadex-LH and eluted with appropriate solvent systems accordingly. In order to obtain Emilia, Italy pure extracts, partially purified fractions were further purified. The structures of the isolated compounds were determined by using data obtained from GC-MS spectrum. The compound Correspondence: Oludare Temitope Osuntokun, Department isolated includeAspidofractinine-3-methanol, Phthalic acid, 2-ethylhexyl tetradecyl of Microbiology, Faculty of Science, Adekunle Ajasin University, ester, Phthalic acid, di (2-propylpentyl) ester), (9-(2’, 2’-Dimethylpropanoilhydrazono)- Akungba-Akoko, Ondo state, Nigeria, 3,6-dichloro-2,7-bis-[2-(diethylamino)-ethoxy]fluorine) and Tere phthalic acid, dodecyl Email 2-ethylhexyl ester.These compounds have individual and synergistic activity against Gram Received: August 08, 2019 | Published: August 26, 2019 negative (E.coli), Gram-positive (Bacillus subtilis) and (Aspergillusflavus) at 10, 5, 2.5 and 1.25 g/mL. -

Correlation and Prediction of Mixing Thermodynamic Properties of Ester-Containing Systems: Ester+Alkane and Ester+Ester Binary S
J. Chem. Thermodynamics 54 (2012) 41–48 Contents lists available at SciVerse ScienceDirect J. Chem. Thermodynamics journal homepage: www.elsevier.com/locate/jct Correlation and prediction of mixing thermodynamic properties of ester-containing systems: Ester + alkane and ester + ester binary systems and the ternary dodecane + ethyl pentanoate + ethyl ethanoate ⇑ Noelia Pérez a, Luís Fernández a, Juan Ortega a, , Francisco J. Toledo a, Jaime Wisniak b a Laboratorio de Termodinámica y Fisicoquímica de Fluidos, Parque Científico-Tecnológico, Universidad de Las Palmas de Gran Canaria, Canary Islands, Spain b Deparment of Chemical Engineering, Ben-Gurion University of the Negev, Beer-Sheva 84105, Israel article info abstract Article history: E E Excess thermodynamic properties V m and Hm, have been measured for the ternary mixture dodecane + Received 25 December 2011 ethyl pentanoate + ethyl ethanoate and for the corresponding binaries dodecane + ethyl pentanoate, Received in revised form 7 March 2012 dodecane + ethyl ethanoate, ethyl pentanoate + ethyl ethanoate at 298.15 K. All mixtures show Accepted 9 March 2012 endothermic and expansive effects. Experimental results are correlated with a suitable equation whose Available online 20 March 2012 final form for the excess ternary quantity ME contains the particular contributions of the three binaries (i–j) and a last term corresponding to the ternary, all of them obtained considering fourth-order interac- Keywords: tions. The fit goodness for all mixtures is good and comparable to others equations taken from the liter- Excess molar enthalpies ature. In this work the dissolution model for the binaries and ternary is analyzed with a special attention Excess molar volumes Ternary mixture to ester–ester binaries whose behaviour is discussed. -

One-Step Synthesis of Ester from Alkene
ー論文ー ●One-step synthesis of ester from alkene Madoka Furuta M.C.Kung and H.H.Kung UFO Project Northwestern University Abstract A one-step synthesis of ethylacetate from a feed mixture of ethene, oxygen, and water using a supported Pd and silicotungstic acid catalyst was demonstrated. At about 180℃ and 25% ethene conversion, ethylacetate could be produced with to 46% selectivity, with 34% acetic acid and ethanol that could be recycled. The catalyst was believed to be bifunctional, with Pd providing the oxidation function and the silicotungstic acid providing the acidic function. And this catalyst system had an activity that could produce isopropylacrylate from a feed mixture of propene, oxygen, and water. 1 Introduction C2H5OH + O2 = CH3COOH + H2O (2) Carboxylic acid esters are used in a wide range of applications. They are used, for example, as plasticizers, solvents, surface-active Finally, ethyl acetate is produced by the esterification of ethanol agents, and flavor and perfume materials. There are many known with acetic acid over a catalyst. methods to produce esters. The most common method is the C2H5OH + CH3COOH = CH3COOC2H5 + H2O (3) esterification, which is a reaction of alcohol with carboxylic acid1,2). For example, ethyl acrylate is synthesized from ethyl alcohol and Thus the overall reaction is acrylic acid, and ethyl acetate from ethyl alcohol and acetic acid. An 2C2H4 + O2 = CH3COOC2H5 (4) acidic catalyst, which can be a mineral acid, or polymeric material containing chemically-bound acid groups, such as sulfonated Such a one-step process has the advantage of starting with reactants, polystyrene, is commonly used in the esterification. -
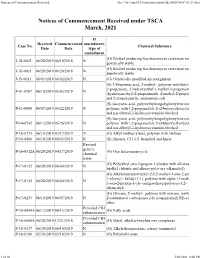
Notices of Commencement Received File:///W:/Oneepa/Newchems/Pubs/5D2-IMD/NOC-03-21.Html
Notices of Commencement Received file:///W:/OneEPA/Newchems/pubs/5d2-IMD/NOC-03-21.html If Received Commencement amendment, Case No. Chemical Substance Date Date type of amendment (G) Biofuel producing Saccharomyces cerevisiae mo J-18-0002 06/28/2019 06/19/2018 N genetically stable (G) Biofuel producing Saccharomyces cerevisiae mo J-18-0003 06/28/2019 06/24/2018 N genetically stable J-19-0011 06/03/2019 06/02/2019 N (G) Genetically modified microorganism (S) 2-Propenoic acid, 2-methyl-, polymer with butyl 2-propenoate, 2-hydroxyethyl 2-methyl-2-propenoat P-01-0767 06/10/2019 05/03/2019 N (hydroxymethyl)-2-propenamide, 2-methyl-2-propen and 2-propenenitrile, ammonium salt (S) Isocyanic acid, polymethylenepolyphenylene este P-11-0080 06/07/2019 05/22/2019 N polymer with 1,2-propanediol, 2-(2-butoxyethoxy)et and n,n-dibutyl-2-hydroxyacetamide-blocked (S) Isocyanic acid, polymethylenepolyphenylene este P-14-0742 06/13/2019 05/26/2019 N polymer with 1,2-propanediol, 2-(2-butoxyethoxy)et and n,n-dibutyl-2-hydroxyacetamide-blocked P-16-0375 06/10/2019 03/17/2019 N (G) Alkyl methacrylates, polymer with olefines P-16-0400 06/18/2019 06/03/2019 N (S) Alkanes, C11-16, branched and linear Revised generic P-16-0532A 06/20/2019 04/17/2019 (G) Oxo heteromonocycle chemical name (G) Poly(alkyl-oxo-2-propen-1-yl)ester with alkanea P-17-0152 06/28/2019 06/04/2019 N trialkyl chloride and alkoxy-poly(oxy-alkanediyl) (G) Alkylaminium-trialkyl-2-[(2-methyl-1-oxo-2-pro 1-yl)oxy]-, halide (1:1), polymer with alpha-(2-meth P-17-0152 06/28/2019 06/04/2019 N 1-oxo-2-propen-1-yl)- -

Ester Synthesis Lab (Student Handout)
Name: ________________________ Lab Partner: ____________________ Date: __________________________ Class Period: ____________________ Ester Synthesis Lab (Student Handout) Lab Report Components: The following must be included in your lab book in order to receive full credit. 1. Purpose 2. Hypothesis 3. Procedure 4. Observation/Data Table 5. Results 6. Mechanism (In class) 7. Conclusion Introduction The compounds you will be making are also naturally occurring compounds; the chemical structure of these compounds is already known from other investigations. Esters are organic molecules of the general form: where R1 and R2 are any carbon chain. Esters are unique in that they often have strong, pleasant odors. As such, they are often used in fragrances, and many artificial flavorings are in fact esters. Esters are produced by the reaction between alcohols and carboxylic acids. For example, reacting ethanol with acetic acid to give ethyl acetate is shown below. + → + In the case of ethyl acetate, R1 is a CH3 group and R2 is a CH3CH2 group. Naming esters systematically requires naming the functional groups on both sides of the bridging oxygen. In the example above, the right side of the ester as shown is a CH3CH2 1 group, or ethyl group. The left side is CH3C=O, or acetate. The name of the ester is therefore ethyl acetate. Deriving the names of the side from the carboxylic acid merely requires replacing the suffix –ic with –ate. Materials • Alcohol • Carboxylic Acid o 1 o A o 2 o B o 3 o C o 4 Observation Parameters: • Record the combination of carboxylic acid and alcohol • Observe each reactant • Observe each product Procedure 1. -
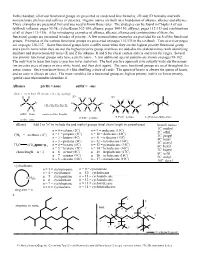
In This Handout, All of Our Functional Groups Are Presented As Condensed Line Formulas, 2D and 3D Formulas and with Nomenclature Prefixes and Suffixes (If Present)
In this handout, all of our functional groups are presented as condensed line formulas, 2D and 3D formulas and with nomenclature prefixes and suffixes (if present). Organic names are built on a foundation of alkanes, alkenes and alkynes. Those examples are presented first and you need to know those rules. The strategies can be found in Chapter 4 of our textbook (alkanes: pages 93-98, cycloalkanes 102-104, alkenes: pages 104-110, alkynes: pages 112-113 and combinations of all of them 113-115). After introducing examples of alkanes, alkenes, alkynes and combinations of them, the functional groups are presented in order of priority. A few nomenclature examples are provided for each of the functional groups. Examples of the various functional groups are presented on pages 115-135 in the textbook. Two overview pages are on pages 136-137. Some functional groups have a suffix name when they are the highest priority functional group and a prefix name when they are not the highest priority group, and these are added to the skeletal names with identifying numbers and stereochemistry terms (E and Z for alkenes, R and S for chiral centers and cis and trans for rings). Several low priority functional groups only have a prefix name. A few additional special patterns are shown on pages 98-102. The only way to learn this topic is practice (over and over). The best practice approach is to actually write out the names (on an extra piece of paper or on a white board, and then do it again). The same functional groups are used throughout the entire course. -

Reactions of Alkenes Since Bonds Are Stronger Than Bonds, Double Bonds Tend to React to Convert the Double Bond Into Bonds
Reactions of Alkenes Since bonds are stronger than bonds, double bonds tend to react to convert the double bond into bonds This is an addition reaction. (Other types of reaction have been substitution and elimination). Addition reactions are typically exothermic. Electrophilic Addition The bond is localized above and below the C-C bond. The electrons are relatively far away from the nuclei and are therefore loosely bound. An electrophile will attract those electrons, and can pull them away to form a new bond. This leaves one carbon with only 3 bonds and a +ve charge (carbocation). The double bond acts as a nucleophile (attacks the electrophile). Ch08 Reacns of Alkenes (landscape) Page 1 In most cases, the cation produced will react with another nucleophile to produce the final overall electrophilic addition product. Electrophilic addition is probably the most common reaction of alkenes. Consider the electrophilic addition of H-Br to but-2-ene: The alkene abstracts a proton from the HBr, and a carbocation and bromide ion are generated. The bromide ion quickly attacks the cationic center and yields the final product. In the final product, H-Br has been added across the double bond. Ch08 Reacns of Alkenes (landscape) Page 2 Orientation of Addition Consider the addition of H-Br to 2-methylbut-2-ene: There are two possible products arising from the two different ways of adding H-Br across the double bond. But only one is observed. The observed product is the one resulting from the more stable carbocation intermediate. Tertiary carbocations are more stable than secondary. -

KEE in Roofing Membranes Only Go So Far – Meaning It Is More Cost-Effective to Use Levels Lower Than Those Required by ASTM D 6754-10
What is KEE? KEE is an abbreviation for Ketone Ethylene Ester, a high-molecular-weight polymer with elastomeric properties and favorable melting properties for thermoplastic processing. Although KEE is most commonly used in roofing, it is also used in geomembranes for secondary containment for petrochemical storage and other industrial chemical applications. It is manufactured solely by DuPont™, under the brand name Elavloy®. There is also an ASTM International standard, ASTM D 6754-10, which regulates the standards for roofing membranes that use KEE. This standard is commonly referred to as the “KEE Standard” and requires that a minimum of 50% by weight of the polymer content of the roofing membrane be KEE. Therefore all roofing membranes that contain KEE may not comply with the “KEE Standard” but still provide many of the same benefits. Why is KEE used in roofing? This molecule can replace plasticizers in roofing membranes as a solution for flexibility that will not migrate out of the membrane. When mixed with PVC, KEE provides many desirable characteristics in roofing membranes, including: Easy heat welding due to melting properties Resistance to chemical attack Weather resistance Long-term flexibility Durability Resistance to microbial growth Energy efficiency What is the difference between ASTM D 4434-12 and ASTM D 6754-10? ASTM D 4434-12* ASTM D 6754-10 This standard was the first of its kind for A newer standard developed to regulate single ply roofing membranes and the polymer content in roofing membranes. provides a nationally-recognized definition This standard requires a minimum content for PVC membranes. It sets benchmarks of 50% KEE, and also has more strict for PVC content, thickness and other requirements for minimum thickness, physical properties applicable to roofing. -

Aldehydes, Ketones and Carboxylic Acids
1212Unit Objectives AldehydesAldehydesAldehydesAldehydes,,,,,, KKKKKKeeeeeetonestonestonestonestonestones After studying this Unit, you will be able to andandandandandand CarboxylicCarboxylicCarboxylicCarboxylicCarboxylicCarboxylic • write the common and IUPAC names of aldehydes, ketones and carboxylic acids; AAAAAAcidscidscidscidscidscids • write the structures of the compounds containing functional groups namely carbonyl and carboxyl groups; Carbonyl compounds are of utmost importance to organic chemistry. They are constituents of fabrics, flavourings, plastics • describe the important methods and drugs. of preparation and reactions of these classes of compounds; In the previous Unit, you have studied organic • correlate physical properties and compounds with functional groups containing carbon- chemical reactions of aldehydes, oxygen single bond. In this Unit, we will study about the ketones and carboxylic acids, organic compounds containing carbon-oxygen double with their structures; bond (>C=O) called carbonyl group, which is one of the • explain the mechanism of a few most important functional groups in organic chemistry. selected reactions of aldehydes and ketones; In aldehydes, the carbonyl group is bonded to a carbon and hydrogen while in the ketones, it is bonded • understand various factors to two carbon atoms. The carbonyl compounds in which affecting the acidity of carboxylic carbon of carbonyl group is bonded to carbon or acids and their reactions; hydrogen and oxygen of hydroxyl moiety (-OH) are • describe the uses of aldehydes, known as carboxylic acids, while in compounds where ketones and carboxylic acids. carbon is attached to carbon or hydrogen and nitrogen of -NH2 moiety or to halogens are called amides and acyl halides respectively. Esters and anhydrides are derivatives of carboxylic acids. The general formulas of these classes of compounds are given below: 2021–22 Aldehydes, ketones and carboxylic acids are widespread in plants and animal kingdom.