Fibrosis Accumulation in Idiopathic Pulmonary Apoptosis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Whole Brain and Brain Regional Coexpression Network Interactions Associated with Predisposition to Alcohol Consumption Lauren A
Virginia Commonwealth University VCU Scholars Compass Study of Biological Complexity Publications Center for the Study of Biological Complexity 2013 Whole Brain and Brain Regional Coexpression Network Interactions Associated with Predisposition to Alcohol Consumption Lauren A. Vanderlinden University of Colorado at Aurora Laura M. Saba University of Colorado at Aurora Katerina Kechris University of Colorado at Aurora See next page for additional authors Follow this and additional works at: http://scholarscompass.vcu.edu/csbc_pubs Part of the Life Sciences Commons © 2013 Vanderlinden et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Downloaded from http://scholarscompass.vcu.edu/csbc_pubs/24 This Article is brought to you for free and open access by the Center for the Study of Biological Complexity at VCU Scholars Compass. It has been accepted for inclusion in Study of Biological Complexity Publications by an authorized administrator of VCU Scholars Compass. For more information, please contact [email protected]. Authors Lauren A. Vanderlinden, Laura M. Saba, Katerina Kechris, Michael F. Miles, Paula L. Hoffman, and Boris Tabakoff This article is available at VCU Scholars Compass: http://scholarscompass.vcu.edu/csbc_pubs/24 Whole Brain and Brain Regional Coexpression Network Interactions Associated with Predisposition to Alcohol Consumption Lauren -

Analysis of Trans Esnps Infers Regulatory Network Architecture
Analysis of trans eSNPs infers regulatory network architecture Anat Kreimer Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2014 © 2014 Anat Kreimer All rights reserved ABSTRACT Analysis of trans eSNPs infers regulatory network architecture Anat Kreimer eSNPs are genetic variants associated with transcript expression levels. The characteristics of such variants highlight their importance and present a unique opportunity for studying gene regulation. eSNPs affect most genes and their cell type specificity can shed light on different processes that are activated in each cell. They can identify functional variants by connecting SNPs that are implicated in disease to a molecular mechanism. Examining eSNPs that are associated with distal genes can provide insights regarding the inference of regulatory networks but also presents challenges due to the high statistical burden of multiple testing. Such association studies allow: simultaneous investigation of many gene expression phenotypes without assuming any prior knowledge and identification of unknown regulators of gene expression while uncovering directionality. This thesis will focus on such distal eSNPs to map regulatory interactions between different loci and expose the architecture of the regulatory network defined by such interactions. We develop novel computational approaches and apply them to genetics-genomics data in human. We go beyond pairwise interactions to define network motifs, including regulatory modules and bi-fan structures, showing them to be prevalent in real data and exposing distinct attributes of such arrangements. We project eSNP associations onto a protein-protein interaction network to expose topological properties of eSNPs and their targets and highlight different modes of distal regulation. -

Proteomic Analysis of the Venom of Jellyfishes Rhopilema Esculentum and Sanderia Malayensis
marine drugs Article Proteomic Analysis of the Venom of Jellyfishes Rhopilema esculentum and Sanderia malayensis 1, 2, 2 2, Thomas C. N. Leung y , Zhe Qu y , Wenyan Nong , Jerome H. L. Hui * and Sai Ming Ngai 1,* 1 State Key Laboratory of Agrobiotechnology, School of Life Sciences, The Chinese University of Hong Kong, Hong Kong, China; [email protected] 2 Simon F.S. Li Marine Science Laboratory, State Key Laboratory of Agrobiotechnology, School of Life Sciences, The Chinese University of Hong Kong, Hong Kong, China; [email protected] (Z.Q.); [email protected] (W.N.) * Correspondence: [email protected] (J.H.L.H.); [email protected] (S.M.N.) Contributed equally. y Received: 27 November 2020; Accepted: 17 December 2020; Published: 18 December 2020 Abstract: Venomics, the study of biological venoms, could potentially provide a new source of therapeutic compounds, yet information on the venoms from marine organisms, including cnidarians (sea anemones, corals, and jellyfish), is limited. This study identified the putative toxins of two species of jellyfish—edible jellyfish Rhopilema esculentum Kishinouye, 1891, also known as flame jellyfish, and Amuska jellyfish Sanderia malayensis Goette, 1886. Utilizing nano-flow liquid chromatography tandem mass spectrometry (nLC–MS/MS), 3000 proteins were identified from the nematocysts in each of the above two jellyfish species. Forty and fifty-one putative toxins were identified in R. esculentum and S. malayensis, respectively, which were further classified into eight toxin families according to their predicted functions. Amongst the identified putative toxins, hemostasis-impairing toxins and proteases were found to be the most dominant members (>60%). -

Regulation of Cdc42 and Its Effectors in Epithelial Morphogenesis Franck Pichaud1,2,*, Rhian F
© 2019. Published by The Company of Biologists Ltd | Journal of Cell Science (2019) 132, jcs217869. doi:10.1242/jcs.217869 REVIEW SUBJECT COLLECTION: ADHESION Regulation of Cdc42 and its effectors in epithelial morphogenesis Franck Pichaud1,2,*, Rhian F. Walther1 and Francisca Nunes de Almeida1 ABSTRACT An overview of Cdc42 Cdc42 – a member of the small Rho GTPase family – regulates cell Cdc42 was discovered in yeast and belongs to a large family of small – polarity across organisms from yeast to humans. It is an essential (20 30 kDa) GTP-binding proteins (Adams et al., 1990; Johnson regulator of polarized morphogenesis in epithelial cells, through and Pringle, 1990). It is part of the Ras-homologous Rho subfamily coordination of apical membrane morphogenesis, lumen formation and of GTPases, of which there are 20 members in humans, including junction maturation. In parallel, work in yeast and Caenorhabditis elegans the RhoA and Rac GTPases, (Hall, 2012). Rho, Rac and Cdc42 has provided important clues as to how this molecular switch can homologues are found in all eukaryotes, except for plants, which do generate and regulate polarity through localized activation or inhibition, not have a clear homologue for Cdc42. Together, the function of and cytoskeleton regulation. Recent studies have revealed how Rho GTPases influences most, if not all, cellular processes. important and complex these regulations can be during epithelial In the early 1990s, seminal work from Alan Hall and his morphogenesis. This complexity is mirrored by the fact that Cdc42 can collaborators identified Rho, Rac and Cdc42 as main regulators of exert its function through many effector proteins. -

Transcriptional Regulation of RKIP in Prostate Cancer Progression
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Sciences Transcriptional Regulation of RKIP in Prostate Cancer Progression Submitted by: Sandra Marie Beach In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Sciences Examination Committee Major Advisor: Kam Yeung, Ph.D. Academic William Maltese, Ph.D. Advisory Committee: Sonia Najjar, Ph.D. Han-Fei Ding, M.D., Ph.D. Manohar Ratnam, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: May 16, 2007 Transcriptional Regulation of RKIP in Prostate Cancer Progression Sandra Beach University of Toledo ACKNOWLDEGMENTS I thank my major advisor, Dr. Kam Yeung, for the opportunity to pursue my degree in his laboratory. I am also indebted to my advisory committee members past and present, Drs. Sonia Najjar, Han-Fei Ding, Manohar Ratnam, James Trempe, and Douglas Pittman for generously and judiciously guiding my studies and sharing reagents and equipment. I owe extended thanks to Dr. William Maltese as a committee member and chairman of my department for supporting my degree progress. The entire Department of Biochemistry and Cancer Biology has been most kind and helpful to me. Drs. Roy Collaco and Hong-Juan Cui have shared their excellent technical and practical advice with me throughout my studies. I thank members of the Yeung laboratory, Dr. Sungdae Park, Hui Hui Tang, Miranda Yeung for their support and collegiality. The data mining studies herein would not have been possible without the helpful advice of Dr. Robert Trumbly. I am also grateful for the exceptional assistance and shared microarray data of Dr. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Toxicogenomics Applications of New Functional Genomics Technologies in Toxicology
\-\w j Toxicogenomics Applications of new functional genomics technologies in toxicology Wilbert H.M. Heijne Proefschrift ter verkrijging vand egraa dva n doctor opgeza gva nd e rector magnificus vanWageninge n Universiteit, Prof.dr.ir. L. Speelman, in netopenbaa r te verdedigen op maandag6 decembe r200 4 des namiddagst e half twee ind eAul a - Table of contents Abstract Chapter I. page 1 General introduction [1] Chapter II page 21 Toxicogenomics of bromobenzene hepatotoxicity: a combined transcriptomics and proteomics approach[2] Chapter III page 48 Bromobenzene-induced hepatotoxicity atth etranscriptom e level PI Chapter IV page 67 Profiles of metabolites and gene expression in rats with chemically induced hepatic necrosis[4] Chapter V page 88 Liver gene expression profiles in relation to subacute toxicity in rats exposed to benzene[5] Chapter VI page 115 Toxicogenomics analysis of liver gene expression in relation to subacute toxicity in rats exposed totrichloroethylen e [6] Chapter VII page 135 Toxicogenomics analysis ofjoin t effects of benzene and trichloroethylene mixtures in rats m Chapter VII page 159 Discussion and conclusions References page 171 Appendices page 187 Samenvatting page 199 Dankwoord About the author Glossary Abbreviations List of genes Chapter I General introduction Parts of this introduction were publishedin : Molecular Biology in Medicinal Chemistry, Heijne etal., 2003 m NATO Advanced Research Workshop proceedings, Heijne eral., 2003 81 Chapter I 1. General introduction 1.1 Background /.1.1 Toxicologicalrisk -

Spatial Protein Interaction Networks of the Intrinsically Disordered Transcription Factor C(%3$
Spatial protein interaction networks of the intrinsically disordered transcription factor C(%3$ Dissertation zur Erlangung des akademischen Grades Doctor rerum naturalium (Dr. rer. nat.) im Fach Biologie/Molekularbiologie eingereicht an der Lebenswissenschaftlichen Fakultät der Humboldt-Universität zu Berlin Von Evelyn Ramberger, M.Sc. Präsidentin der Humboldt-Universität zu Berlin Prof. Dr.-Ing.Dr. Sabine Kunst Dekan der Lebenswissenschaftlichen Fakultät der Humboldt-Universität zu Berlin Prof. Dr. Bernhard Grimm Gutachter: 1. Prof. Dr. Achim Leutz 2. Prof. Dr. Matthias Selbach 3. Prof. Dr. Gunnar Dittmar Tag der mündlichen Prüfung: 12.8.2020 For T. Table of Contents Selbstständigkeitserklärung ....................................................................................1 List of Figures ............................................................................................................2 List of Tables ..............................................................................................................3 Abbreviations .............................................................................................................4 Zusammenfassung ....................................................................................................6 Summary ....................................................................................................................7 1. Introduction ............................................................................................................8 1.1. Disordered proteins -

High Throughput Strategies Aimed at Closing the GAP in Our Knowledge of Rho Gtpase Signaling
cells Review High Throughput strategies Aimed at Closing the GAP in Our Knowledge of Rho GTPase Signaling Manel Dahmene 1, Laura Quirion 2 and Mélanie Laurin 1,3,* 1 Oncology Division, CHU de Québec–Université Laval Research Center, Québec, QC G1V 4G2, Canada; [email protected] 2 Montréal Clinical Research Institute (IRCM), Montréal, QC H2W 1R7, Canada; [email protected] 3 Université Laval Cancer Research Center, Québec, QC G1R 3S3, Canada * Correspondence: [email protected] Received: 21 May 2020; Accepted: 7 June 2020; Published: 9 June 2020 Abstract: Since their discovery, Rho GTPases have emerged as key regulators of cytoskeletal dynamics. In humans, there are 20 Rho GTPases and more than 150 regulators that belong to the RhoGEF, RhoGAP, and RhoGDI families. Throughout development, Rho GTPases choregraph a plethora of cellular processes essential for cellular migration, cell–cell junctions, and cell polarity assembly. Rho GTPases are also significant mediators of cancer cell invasion. Nevertheless, to date only a few molecules from these intricate signaling networks have been studied in depth, which has prevented appreciation for the full scope of Rho GTPases’ biological functions. Given the large complexity involved, system level studies are required to fully grasp the extent of their biological roles and regulation. Recently, several groups have tackled this challenge by using proteomic approaches to map the full repertoire of Rho GTPases and Rho regulators protein interactions. These studies have provided in-depth understanding of Rho regulators specificity and have contributed to expand Rho GTPases’ effector portfolio. Additionally, new roles for understudied family members were unraveled using high throughput screening strategies using cell culture models and mouse embryos. -

Integrating Protein Copy Numbers with Interaction Networks to Quantify Stoichiometry in Mammalian Endocytosis
bioRxiv preprint doi: https://doi.org/10.1101/2020.10.29.361196; this version posted October 29, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Integrating protein copy numbers with interaction networks to quantify stoichiometry in mammalian endocytosis Daisy Duan1, Meretta Hanson1, David O. Holland2, Margaret E Johnson1* 1TC Jenkins Department of Biophysics, Johns Hopkins University, 3400 N Charles St, Baltimore, MD 21218. 2NIH, Bethesda, MD, 20892. *Corresponding Author: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/2020.10.29.361196; this version posted October 29, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Abstract Proteins that drive processes like clathrin-mediated endocytosis (CME) are expressed at various copy numbers within a cell, from hundreds (e.g. auxilin) to millions (e.g. clathrin). Between cell types with identical genomes, copy numbers further vary significantly both in absolute and relative abundance. These variations contain essential information about each protein’s function, but how significant are these variations and how can they be quantified to infer useful functional behavior? Here, we address this by quantifying the stoichiometry of proteins involved in the CME network. We find robust trends across three cell types in proteins that are sub- vs super-stoichiometric in terms of protein function, network topology (e.g. -

Atlas Journal
Atlas of Genetics and Cytogenetics in Oncology and Haematology Home Genes Leukemias Solid Tumours Cancer-Prone Deep Insight Portal Teaching X Y 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 NA Atlas Journal Atlas Journal versus Atlas Database: the accumulation of the issues of the Journal constitutes the body of the Database/Text-Book. TABLE OF CONTENTS Volume 5, Number 2, Apr-Jun 2001 Previous Issue / Next Issue Genes COL1A1 (collagen, type I, alpha 1) (17q21.31-q22). Marie-Pierre Simon, Georges Maire, Florence Pedeutour. Atlas Genet Cytogenet Oncol Haematol 2001; 5 (2): 169-177. [Full Text] [PDF] URL : http://AtlasGeneticsOncology.org/Genes/COL1A1ID186.html NF2 (neurofibromatosis type 2) (22q12.1-12.2) - updated. James F Gusella. Atlas Genet Cytogenet Oncol Haematol 2001; 5 (2): 178-184. [Full Text] [PDF] URL : http://AtlasGeneticsOncology.org/Genes/NF2117.html PDGFB (22q12.3-q13.1). Marie-Pierre Simon, Georges Maire, Florence Pedeutour. Atlas Genet Cytogenet Oncol Haematol 2001; 5 (2): 185-194. [Full Text] [PDF] URL : http://AtlasGeneticsOncology.org/Genes/PDGFBID155.html XPA (9q22.3-9q22.3). Anne Stary, Alain Sarasin. Atlas Genet Cytogenet Oncol Haematol 2001; 5 (2): 195-204. [Full Text] [PDF] Atlas Genet Cytogenet Oncol Haematol 2001; 2 I URL : http://AtlasGeneticsOncology.org/Genes/XPAID104.html ERCC-3 (Excision repair cross-complementing rodent repair deficiency, complementation group 3) (2q21). Anne Stary, Alain Sarasin. Atlas Genet Cytogenet Oncol Haematol 2001; 5 (2): 205-214. [Full Text] [PDF] URL : http://AtlasGeneticsOncology.org/Genes/XPBID296.html XPC (3p25.1). -
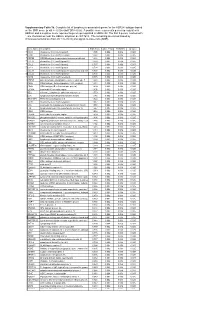
* Supplementary Table 3B. Complete List of Lymphocytic-Associated Genes
Supplementary Table 3b. Complete list of lymphocytic-associated genes for the HER2+I subtype based on the SNR score (p-val <= 0.005 and FDR<=0.05). A positive score represents genes up-regulated in HER2+I and a negative score represents genes up-regulated in HER2+NI. The first 9 genes, marked with *, are chemokines near the HER2+ amplicon at chr17q12. The remaining are sorted based by chromosomal location (from chr 1 to chr X) and signal-to-noise ratio (SNR). Gene Name Description SNR Score Feature P value FDR(BH) Q Value * CCL5 chemokine (C-C motif) ligand 5 1.395 0.002 0.036 0.025 * CCR7 chemokine (C-C motif) receptor 7 1.362 0.002 0.036 0.025 * CD79B CD79B antigen (immunoglobulin-associated beta) 1.248 0.002 0.036 0.025 * CCL13 chemokine (C-C motif) ligand 13 1.003 0.002 0.036 0.025 * CCL2 chemokine (C-C motif) ligand 2 0.737 0.004 0.056 0.037 * CCL8 chemokine (C-C motif) ligand 8 0.724 0.002 0.036 0.025 * CCL18 chemokine (C-C motif) ligand 18 (pulmonary and activ 0.703 0.002 0.036 0.025 * CCL23 chemokine (C-C motif) ligand 23 0.701 0.002 0.036 0.025 * CCR6 chemokine (C-C motif) receptor 6 0.715 0.002 0.036 0.025 PTPN7 protein tyrosine phosphatase, non-receptor type 7 1.861 0.002 0.036 0.025 CD3Z CD3Z antigen, zeta polypeptide (TiT3 complex) 1.811 0.002 0.036 0.025 CD48 CD48 antigen (B-cell membrane protein) 1.809 0.002 0.036 0.025 IL10RA interleukin 10 receptor, alpha 1.806 0.002 0.036 0.025 SELL selectin L (lymphocyte adhesion molecule 1) 1.773 0.002 0.036 0.025 LCK lymphocyte-specific protein tyrosine kinase 1.745 0.002 0.036 0.025