ACVR2B Recombinant Protein Cat
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Lack of Tgfbr1 and Acvr1b Synergistically Stimulates Myofibre Hypertrophy And
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.03.433740; this version posted March 6, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Lack of Tgfbr1 and Acvr1b synergistically stimulates myofibre hypertrophy and accelerates muscle regeneration *M.M.G. Hillege1, *A. Shi1,2 ,3, R.C. Galli Caro1, G. Wu4, P. Bertolino5, W.M.H. Hoogaars1,6, R.T. Jaspers1 1. Laboratory for Myology, Department of Human Movement Sciences, Faculty of Behavioural and Movement Sciences, Vrije Universiteit Amsterdam, Amsterdam Movement Sciences, Amsterdam, The Netherlands 2. Department of Oral and Maxillofacial Surgery/Pathology, Amsterdam UMC and Academic Center for Dentistry Amsterdam (ACTA), Vrije Universiteit Amsterdam (VU), Amsterdam Movement Sciences (AMS), Amsterdam, the Netherlands 3. Key Laboratory of Oral Medicine, Guangzhou Institute of Oral Disease, Affiliated Stomatology Hospital of Guangzhou Medical University, Guangzhou Medical University, Guangzhou, China 4. Department of Oral Implantology and Prosthetic Dentistry, Academic Centre for Dentistry Amsterdam (ACTA), University of Amsterdam (UvA) and Vrije Universiteit Amsterdam (VU), The Netherlands 5. Centre de Recherche en Cancérologie de Lyon, UMR INSERM U1052/CNRS 5286, Université de Lyon, Centre Léon Bérard, Lyon, France 6. European Research Institute for the Biology of Ageing (ERIBA), University Medical Center Groningen (UMCG), University of Groningen, Groningen, The Netherlands *Contributed equally to this manuscript **Correspondence: [email protected]; Tel.: +31 (0) 205988463 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.03.03.433740; this version posted March 6, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. -

In Order to Measure the Binding Constants of ACVR1 Mabs, Mabs Were Captured with an Anti-Human Fc Antibody Immobilized on a CM5 Chip
Supplemental Table 1: Binding constants of ACVR1 Mabs and Fabs to human ACVR1 1/2 ACVR1 Mab ka kd KD t Tested (1/Ms) (1/s) (M) (min) Mab 1 1.31E+06 1.59E-03 1.21E-09 7 Mab 2 7.18E+05 1.63E-04 2.27E-10 71 Mab 3 6.77E+05 1.76E-04 2.60E-10 65 Fab 2 5.49E+05 9.36E-05 1.70E-10 123 Fab 3 6.90E+05 1.06E-04 1.54E-10 109 hACVR1 2.30E+05 1.33E-03 5.80E-09 9 Mab In order to measure the binding constants of ACVR1 Mabs, Mabs were captured with an anti-human Fc antibody immobilized on a CM5 chip. Different concentrations of hACVR1.mmh (REGN3111) were injected over ACVR1 Mabs at 37 0C. In order to measure the binding constants of ACVR1 Fabs, hACVR1.mmh was captured with a myc antibody (REGN642) immobilized on a CM5 chip. Different concentrations of ACVR1 Fabs were injected over hACVR1.mmh at 37 0C. Binding rate constants and equilibrium dissociation rate constants were calculated by fitting data using 1:1 Langmuir binding model (Scrubber 2.0c). All 3 ACVR1 Fabs bound to monomeric human ACVR1 with binding kinetics similar (< 2.5-fold difference) to their respective Mabs. Supplemental Table 2: Binding constants of ACVR1 Mabs and Fabs to mouse ACVR1 1/2 ACVR1 Mab ka kd KD t Tested (1/Ms) (1/s) (M) (min) Mab 1 1.34E+06 1.67E-03 1.24E-09 7 Mab 2 7.13E+05 1.61E-04 2.26E-10 72 Mab 3 6.74E+05 1.81E-04 2.68E-10 64 Fab 2 5.45E+05 9.72E-05 1.78E-10 119 Fab 3 6.53E+05 1.05E-04 1.60E-10 110 hACVR1 ND ND ND ND Mab In order to measure the binding constants of ACVR1 Mabs, Mabs were captured with an anti-human Fc antibody immobilized on a CM5 chip. -

Targeting Myostatin/Activin a Protects Against Skeletal Muscle and Bone Loss During Spaceflight
Targeting myostatin/activin A protects against skeletal muscle and bone loss during spaceflight Se-Jin Leea,b,1, Adam Lehara, Jessica U. Meirc, Christina Kochc, Andrew Morganc, Lara E. Warrend, Renata Rydzike, Daniel W. Youngstrome, Harshpreet Chandoka, Joshy Georgea, Joseph Gogainf, Michael Michauda, Thomas A. Stoklaseka, Yewei Liua, and Emily L. Germain-Leeg,h aThe Jackson Laboratory for Genomic Medicine, Farmington, CT 06032; bDepartment of Genetics and Genome Sciences, University of Connecticut School of Medicine, Farmington, CT 06030; cThe National Aeronautics and Space Administration, NASA Johnson Space Center, Houston, TX 77058; dCenter for the Advancement of Science in Space, Houston, TX 77058; eDepartment of Orthopaedic Surgery, University of Connecticut School of Medicine, Farmington, CT 06030; fSomaLogic, Inc., Boulder, CO 80301; gDepartment of Pediatrics, University of Connecticut School of Medicine, Farmington, CT 06030; and hConnecticut Children’s Center for Rare Bone Disorders, Farmington, CT 06032 Contributed by Se-Jin Lee, August 4, 2020 (sent for review July 14, 2020; reviewed by Shalender Bhasin and Paul Gregorevic) Among the physiological consequences of extended spaceflight signaling and, as a result, can induce significant muscle growth are loss of skeletal muscle and bone mass. One signaling pathway when given systemically to wild type mice (13). Indeed, by that plays an important role in maintaining muscle and bone blocking both ligands, this decoy receptor can induce signifi- homeostasis is that regulated by the secreted signaling proteins, cantly more muscle growth than other MSTN inhibitors, and at myostatin (MSTN) and activin A. Here, we used both genetic and high doses, ACVR2B/Fc can induce over 50% muscle growth in pharmacological approaches to investigate the effect of targeting just 2 wk. -

Cardiovascular Diseases Genetic Testing Program Information
Cardiovascular Diseases Genetic Testing Program Description: Congenital Heart Disease Panels We offer comprehensive gene panels designed to • Congenital Heart Disease Panel (187 genes) diagnose the most common genetic causes of hereditary • Heterotaxy Panel (114 genes) cardiovascular diseases. Testing is available for congenital • RASopathy/Noonan Spectrum Disorders Panel heart malformation, cardiomyopathy, arrythmia, thoracic (31 genes) aortic aneurysm, pulmonary arterial hypertension, Marfan Other Panels syndrome, and RASopathy/Noonan spectrum disorders. • Pulmonary Arterial Hypertension (PAH) Panel Hereditary cardiovascular disease is caused by variants in (20 genes) many different genes, and may be inherited in an autosomal dominant, autosomal recessive, or X-linked manner. Other Indications: than condition-specific panels, we also offer single gene Panels: sequencing for any gene on the panels, targeted variant • Confirmation of genetic diagnosis in a patient with analysis, and targeted deletion/duplication analysis. a clinical diagnosis of cardiovascular disease Tests Offered: • Carrier or pre-symptomatic diagnosis identification Arrythmia Panels in individuals with a family history of cardiovascular • Comprehensive Arrhythmia Panel (81 genes) disease of unknown genetic basis • Atrial Fibrillation (A Fib) Panel (28 genes) Gene Specific Sequencing: • Atrioventricular Block (AV Block) Panel (7 genes) • Confirmation of genetic diagnosis in a patient with • Brugada Syndrome Panel (21 genes) cardiovascular disease and in whom a specific -

Functional Redundancy of Type I and Type II Receptors in the Regulation Of
Functional redundancy of type I and type II receptors in INAUGURAL ARTICLE the regulation of skeletal muscle growth by myostatin and activin A Se-Jin Leea,b,1, Adam Lehara, Yewei Liua, Chi Hai Lyc,d, Quynh-Mai Phama, Michael Michauda, Renata Rydzike, Daniel W. Youngstrome, Michael M. Shenf, Vesa Kaartineng, Emily L. Germain-Leeh,i, and Thomas A. Randoc,d,j aThe Jackson Laboratory for Genomic Medicine, Farmington, CT 06032; bDepartment of Genetics and Genome Sciences, University of Connecticut School of Medicine, Farmington, CT 06030; cPaul F. Glenn Center for the Biology of Aging, Stanford University School of Medicine, Stanford, CA 94305; dDepartment of Neurology and Neurological Sciences, Stanford University School of Medicine, Stanford, CA 94305; eDepartment of Orthopaedic Surgery, University of Connecticut School of Medicine, Farmington, CT 06030; fDepartment of Genetics and Development, Columbia University, New York, NY 10032; gDepartment of Biologic and Materials Sciences and Prosthodontics, University of Michigan School of Dentistry, Ann Arbor, MI 48109; hDepartment of Pediatrics, University of Connecticut School of Medicine, Farmington, CT 06030; iConnecticut Children’s Center for Rare Bone Disorders, Farmington, CT 06032; and jNeurology Service, VA Palo Alto Health Care System, Palo Alto, CA 94304 This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected in 2012. Contributed by Se-Jin Lee, October 13, 2020 (sent for review September 14, 2020; reviewed by Chen-Ming Fan and S. Paul Oh) Myostatin (MSTN) is a transforming growth factor-β (TGF-β) family atrophy following falls and hip fracture surgery, age-related member that normally acts to limit muscle growth. -

Characterization of Tgfβ-Associated Molecular Features and Drug
Zhang et al. BMC Gastroenterol (2021) 21:284 https://doi.org/10.1186/s12876-021-01869-4 RESEARCH Open Access Characterization of TGFβ-associated molecular features and drug responses in gastrointestinal adenocarcinoma Qiaofeng Zhang1,2,3†, Furong Liu1,2,3†, Lu Qin4, Zhibin Liao1,2,3, Jia Song1,2,3, Huifang Liang1,2,3, Xiaoping Chen1,2,3, Zhanguo Zhang1,2,3* and Bixiang Zhang1,2,3* Abstract Background: Gastrointestinal adenocarcinoma (GIAD) has caused a serious disease burden globally. Targeted ther- apy for the transforming growth factor beta (TGF-β) signaling pathway is becoming a reality. However, the molecular characterization of TGF-β associated signatures in GIAD requires further exploration. Methods: Multi-omics data were collected from TCGA and GEO database. A pivotal unsupervised clustering for TGF-β level was performed by distinguish status of TGF-β associated genes. We analyzed diferential mRNAs, miRNAs, proteins gene mutations and copy number variations in both clusters for comparison. Enrichment of pathways and gene sets were identifed in each type of GIAD. Then we performed diferential mRNA related drug response by col- lecting data from GDSC. At last, a summarized deep neural network for TGF-β status and GIADs was constracted. Results: The TGF-βhigh group had a worse prognosis in overall GIAD patients, and had a worse prognosis trend in gastric cancer and colon cancer specifcally. Signatures (including mRNA and proteins) of the TGF-βhigh group is highly correlated with EMT. According to miRNA analysis, miR-215-3p, miR-378a-5p, and miR-194-3p may block the efect of TGF-β. -

Signaling Receptors for TGF-B Family Members
Downloaded from http://cshperspectives.cshlp.org/ on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press Signaling Receptors for TGF-b Family Members Carl-Henrik Heldin1 and Aristidis Moustakas1,2 1Ludwig Institute for Cancer Research Ltd., Science for Life Laboratory, Uppsala University, SE-751 24 Uppsala, Sweden 2Department of Medical Biochemistry and Microbiology, Science for Life Laboratory, Uppsala University, SE-751 23 Uppsala, Sweden Correspondence: [email protected] Transforming growth factor b (TGF-b) family members signal via heterotetrameric complexes of type I and type II dual specificity kinase receptors. The activation and stability of the receptors are controlled by posttranslational modifications, such as phosphorylation, ubiq- uitylation, sumoylation, and neddylation, as well as by interaction with other proteins at the cell surface and in the cytoplasm. Activation of TGF-b receptors induces signaling via formation of Smad complexes that are translocated to the nucleus where they act as tran- scription factors, as well as via non-Smad pathways, including the Erk1/2, JNK and p38 MAP kinase pathways, and the Src tyrosine kinase, phosphatidylinositol 30-kinase, and Rho GTPases. he transforming growth factor b (TGF-b) embryonic development and in the regulation Tfamily of cytokine genes has 33 human of tissue homeostasis, through their abilities to members, encoding TGF-b isoforms, bone regulate cell proliferation, migration, and differ- morphogenetic proteins (BMPs), growth and entiation. Perturbation of signaling by TGF-b differentiation factors (GDFs), activins, inhib- family members is often seen in different dis- ins, nodal, and anti-Mu¨llerian hormone (AMH) eases, including malignancies, inflammatory (Derynck and Miyazono 2008; Moustakas and conditions, and fibrotic conditions. -

Mirna Replacement in Aortic Calcification Ying Tang1, †, Tapan A
MicroRNA profiles in calcified and healthy aorta: therapeutic impact of miR-145 and miR-378 Running title: miRNA replacement in aortic calcification Ying Tang1, †, Tapan A. Shah1, †, ‡, Edward J. Yurkow2, and Melissa B. Rogers1, ‡ 1Rutgers - New Jersey Medical School, Microbiology, Biochemistry, & Molecular Genetics, Newark, NJ 2Rutgers University Molecular Imaging Center (RUMIC), Rutgers University, Piscataway, NJ †Co-first authors ‡Present Address: Advanced Cell Diagnostics, 7707 Gateway Blvd #200, Newark, CA 94560 §To whom should correspondence and reprint request be addressed to: Melissa B. Rogers, Ph.D., Microbiology, Biochemistry & Molecular Genetics, Rutgers - NJ Medical School (NJMS), Center for Cell Signaling, Room F1216, 205 South Orange Ave., Newark, NJ 07103. Email: [email protected], telephone: 973 972 2984 Fig. S1. Experimental Controls. Fig. S2. KEGG (Kyoto Encyclopedia of Genes and Genomes) analyses of differentially expressed miRNAs in male Klotho homozygous mutant mice. Fig. S3. KEGG (Kyoto Encyclopedia of Genes and Genomes) analyses of differentially expressed miRNAs in female Klotho homozygous mutant mice. Fig. S4. Gene Ontology (GO) analyses of differentially expressed miRNAs in male Klotho homozygous mutant mice. Fig. S5. Gene Ontology (GO) analyses of differentially expressed miRNAs in female Klotho homozygous mutant mice. Fig. S6. Interaction network of miRNAs down-regulated in male Klotho homozygotes relative to healthy controls. Fig. S7. Interaction network of miRNAs down-regulated in female Klotho homozygotes relative to healthy controls. Table S1. Klotho wild type and Klotho heterozygous mice are equivalent controls. Table S2. Average miRNA abundance in aorta from Klotho mutant homozygotes relative to healthy control (p<0.05). Table S3. Genes targeted by selected miRNAs that may influence aortic calcification. -

BMPR2 Inhibits Activin- and BMP-Signaling Via Wild Type ALK2
bioRxiv preprint doi: https://doi.org/10.1101/222406; this version posted November 22, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 BMPR2 inhibits activin- and BMP-signaling via wild type ALK2 2 3 Oddrun Elise Olsen1,2, Meenu Sankar3, Samah Elsaadi1, Hanne Hella1, Glenn Buene1, Sagar 4 Ramesh Darvekar1, Kristine Misund1,2, Takenobu Katagiri4 and Toril Holien1,2* 5 6 (1) Department of Clinical and Molecular Medicine, NTNU – Norwegian University of 7 Science and Technology, Trondheim, Norway. 8 (2) Clinic of Medicine, St. Olav’s University Hospital, Trondheim, Norway. 9 (3) School of Bioscience, University of Skövde, Skövde, Sweden. 10 (4) Division of Pathophysiology, Research Center for Genomic Medicine, Saitama Medical 11 University, Hidaka-shi, Saitama 350-1241, Japan. 12 13 * Corresponding author 14 E-mail: [email protected] 15 16 17 Running title: BMPR2 inhibits ALK2 activity 18 19 Key words: Bone Morphogenetic Protein, BMPR2, ACVR2A, ACVR2B, Activin, ACVR1 1 bioRxiv preprint doi: https://doi.org/10.1101/222406; this version posted November 22, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Summary Statement 2 The activation of SMAD1/5/8 via endogenous wild type ALK2 by activin A, activin B, and 3 certain BMPs was enhanced when BMPR2 levels were knocked down. 4 5 6 Abstract 7 Activin A is a member of the TGF-β superfamily and activates the transcription factors 8 SMAD2/3 through the ALK4 type 1 receptor. -

Targeting Cancer with Kinase Inhibitors
Targeting cancer with kinase inhibitors Stefan Gross, … , Christoph Lengauer, Klaus P. Hoeflich J Clin Invest. 2015;125(5):1780-1789. https://doi.org/10.1172/JCI76094. Review Kinase inhibitors have played an increasingly prominent role in the treatment of cancer and other diseases. Currently, more than 25 oncology drugs that target kinases have been approved, and numerous additional therapeutics are in various stages of clinical evaluation. In this Review, we provide an in-depth analysis of activation mechanisms for kinases in cancer, highlight recent successes in drug discovery, and demonstrate the clinical impact of selective kinase inhibitors. We also describe the substantial progress that has been made in designing next-generation inhibitors to circumvent on- target resistance mechanisms, as well as ongoing strategies for combining kinase inhibitors in the clinic. Last, there are numerous prospects for the discovery of novel kinase targets, and we explore cancer immunotherapy as a new and promising research area for studying kinase biology. Find the latest version: https://jci.me/76094/pdf REVIEW The Journal of Clinical Investigation Targeting cancer with kinase inhibitors Stefan Gross, Rami Rahal, Nicolas Stransky, Christoph Lengauer, and Klaus P. Hoeflich Blueprint Medicines, Cambridge, Massachusetts, USA. Kinase inhibitors have played an increasingly prominent role in the treatment of cancer and other diseases. Currently, more than 25 oncology drugs that target kinases have been approved, and numerous additional therapeutics are in various stages of clinical evaluation. In this Review, we provide an in-depth analysis of activation mechanisms for kinases in cancer, highlight recent successes in drug discovery, and demonstrate the clinical impact of selective kinase inhibitors. -

1 ACVR1 Antibodies Exacerbate Heterotopic Ossification In
bioRxiv preprint doi: https://doi.org/10.1101/2021.07.18.452865; this version posted July 19, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 ACVR1 antibodies exacerbate heterotopic ossification in fibrodysplasia ossificans progressiva (FOP) by activating FOP-mutant ACVR1 Senem Aykul1* Lily Huang1* Lili Wang1 Nanditha Das1 Sandra Reisman1 Yonaton Ray1 Qian Zhang1 Nyanza Rothman1 Kalyan C. Nannuru1 Vishal Kamat1 Susannah Brydges1 Luca Troncone2 Laura Johnsen1 Paul B. Yu2 John Lees-Shepard1 Kevin Schutz3 Andrew J. Murphy1 Aris N. Economides1† Vincent Idone1† Sarah J. Hatsell1† *Indicates equal contribution. †Indicates equal contribution; to whom correspondence should be addressed. [email protected], [email protected], [email protected] 1Regeneron Pharmaceuticals Inc, 777 Old Saw Mill River Road, Tarrytown, NY 10591, USA. 2Department of Medicine, Cardiovascular Division, Brigham and Women's Hospital, Harvard Medical School, Boston, MA 02114, USA. 3Adimab, 7 Lucent Dr, Lebanon, NH 03766, USA. bioRxiv preprint doi: https://doi.org/10.1101/2021.07.18.452865; this version posted July 19, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 2 Abstract Fibrodysplasia ossificans progressiva (FOP) is a rare genetic disorder whose most debilitating pathology is progressive and cumulative heterotopic ossification (HO) of skeletal muscles, ligaments, tendons, and fascia. -
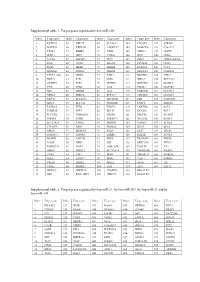
Target Genes Regulated by Hsa-Mir-21, by Hsa-Mir-203, by Hsa-Mir-21 and by Hsa-Mir-143
Supplemental table 1: Target genes regulated by hsa-miR-205 Index Target gene Index Target gene Index Target gene Index Target gene Index Target gene 1 KCTD20 35 UBE2Z 69 SLC38A1 103 LPCAT1 137 STK38L 2 MAPK14 36 YWHAH 70 ANGPTL7 104 MARCKS 138 C1orf123 3 TXNL1 37 RBBP4 71 CTGF 105 MED13 139 GUCD1 4 SPDL1 38 LRP1 72 CYR61 106 IPO7 140 CDK6 5 TCF20 39 IMPAD1 73 TP73 107 PHC2 141 CDKN2AIPNL 6 RAN 40 GNAS 74 EGLN2 108 PICALM 142 CLIP1 7 RGS6 41 MED1 75 ERBB2 109 PLAGL2 143 CUL5 8 HOXA11 42 INPPL1 76 PRRG4 110 NDUFA4 144 C6orf201 9 PAPPA-AS1 43 DDX5 77 F2RL2 111 NDUFB2 145 VTI1A 10 PRR15 44 E2F1 78 GOT1 112 NIPA2 146 SLC5A12 11 ACTRT3 45 E2F5 79 NUFIP2 113 NOTCH2 147 MAML2 12 YES1 46 ZEB2 80 IL24 114 PANK1 148 MAP3K9 13 SRC 47 ERBB3 81 IL32 115 PARD6B 149 NUDT21 14 NPRL3 48 PRKCE 82 RNF217 116 TMEM66 150 DNAJA1 15 NFAT5 49 SLC41A1 83 ZNF585B 117 EZR 151 CCDC108 16 XPOT 50 SLC7A2 84 SIGMAR1 118 ENPP4 152 SHISA6 17 KCTD16 51 ZEB1 85 VEGFA 119 LRRTM4 153 ACP1 18 TMSB4X 52 PHF8 86 BCL9L 120 KCNJ10 154 BCL2 19 PLCXD2 53 TMEM201 87 CREB1 121 PHLPP2 155 NCAPG 20 TNFSF8 54 PTPRJ 88 SERINC3 122 YEATS2 156 KLHL5 21 SLC25A25 55 ETNK1 89 HMGB3 123 VAMP1 157 ACSL4 22 C11orf74 56 XPR1 90 SRD5A1 124 RTN3 158 BCL6 23 GM2A 57 MRPL44 91 PTEN 125 RFX7 159 ITGA5 24 SMNDC1 58 TM9SF2 92 ESRRG 126 RAP2B 160 ACSL1 25 BAMBI 59 PAIP2B 93 PRLR 127 TRAF3IP1 161 EID2B 26 LCOR 60 NEK9 94 ICK 128 SERTAD2 162 TEX35 27 TMEM239 61 NOX5 95 LOH12CR1 129 TOLLIP 163 YY1 28 AMOT 62 DMXL2 96 SLC39A14 130 TMEM55B 164 SMAD1 29 CDK1 63 ETF1 97 BDP1 131 TMEM123 165 SMAD4 30 SQLE 64