The Transcriptomic Architecture of Mouse Sertoli Cell Clone Embryos Reveals Temporal–Spatial-Specific Reprogramming
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Membrane Tension Buffering by Caveolae: a Role in Cancer?
Cancer and Metastasis Reviews (2020) 39:505–517 https://doi.org/10.1007/s10555-020-09899-2 Membrane tension buffering by caveolae: a role in cancer? Vibha Singh1 & Christophe Lamaze1 Published online: 30 May 2020 # Springer Science+Business Media, LLC, part of Springer Nature 2020 Abstract Caveolae are bulb-like invaginations made up of two essential structural proteins, caveolin-1 and cavins, which are abundantly present at the plasma membrane of vertebrate cells. Since their discovery more than 60 years ago, the function of caveolae has been mired in controversy. The last decade has seen the characterization of new caveolae components and regulators together with the discovery of additional cellular functions that have shed new light on these enigmatic structures. Early on, caveolae and/ or caveolin-1 have been involved in the regulation of several parameters associated with cancer progression such as cell migration, metastasis, angiogenesis, or cell growth. These studies have revealed that caveolin-1 and more recently cavin-1 have a dual role with either a negative or a positive effect on most of these parameters. The recent discovery that caveolae can act as mechanosensors has sparked an array of new studies that have addressed the mechanobiology of caveolae in various cellular functions. This review summarizes the current knowledge on caveolae and their role in cancer development through their activity in membrane tension buffering. We propose that the role of caveolae in cancer has to be revisited through their response to the mechanical forces encountered by cancer cells during tumor mass development. Keywords Caveolae . Cancer . Mechanosensing . Mechanotransdcution . Membrane tension . -

Identification of Key Pathways and Genes in Endometrial Cancer Using Bioinformatics Analyses
ONCOLOGY LETTERS 17: 897-906, 2019 Identification of key pathways and genes in endometrial cancer using bioinformatics analyses YAN LIU, TENG HUA, SHUQI CHI and HONGBO WANG Department of Obstetrics and Gynecology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei 430022, P.R. China Received March 16, 2018; Accepted October 12, 2018 DOI: 10.3892/ol.2018.9667 Abstract. Endometrial cancer (EC) is one of the most Introduction common gynecological cancer types worldwide. However, to the best of our knowledge, its underlying mechanisms Endometrial carcinoma (EC) is one of the most common remain unknown. The current study downloaded three mRNA gynecological cancer types, with increasing global incidence and microRNA (miRNA) datasets of EC and normal tissue in recent years (1). A total of 60,050 cases of EC and 10,470 samples, GSE17025, GSE63678 and GSE35794, from the EC-associated cases of mortality were reported in the USA in Gene Expression Omnibus to identify differentially expressed 2016 (1), which was markedly higher than the 2012 statistics genes (DEGs) and miRNAs (DEMs) in EC tumor tissues. of 47,130 cases and 8,010 mortalities (2). Although numerous The DEGs and DEMs were then validated using data from studies have been conducted to investigate the mechanisms of The Cancer Genome Atlas and subjected to gene ontology endometrial tumorigenesis and development, to the best of our and Kyoto Encyclopedia of Genes and Genomes pathway knowledge, the exact etiology remains unknown. Understanding analysis. STRING and Cytoscape were used to construct a the potential molecular mechanisms underlying EC initiation protein-protein interaction network and the prognostic effects and progression is of great clinical significance. -

EHD2 Is a Mechanotransducer Connecting Caveolae Dynamics with Gene Transcription
EHD2 is a mechanotransducer connecting caveolae dynamics with gene transcription Satish Kailasam Mani, Cesar Valades-Cruz, Stéphanie Torrino, Wei-Wei Shen, Cedric Blouin, Satish Kailasam Mani, Christine Viaris de Lesegno, Pierre Bost, Alexandre Grassart, Darius Köster, et al. To cite this version: Satish Kailasam Mani, Cesar Valades-Cruz, Stéphanie Torrino, Wei-Wei Shen, Cedric Blouin, et al.. EHD2 is a mechanotransducer connecting caveolae dynamics with gene transcription. Journal of Cell Biology, Rockefeller University Press, 2018, 217 (12), pp.4092-4105. 10.1083/jcb.201801122. inserm- 02426440 HAL Id: inserm-02426440 https://www.hal.inserm.fr/inserm-02426440 Submitted on 2 Jan 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution - NonCommercial - ShareAlike| 4.0 International License REPORT EHD2 is a mechanotransducer connecting caveolae dynamics with gene transcription Stéphanie Torrino1,2,3*, Wei‑Wei Shen1,2,3*, Cédric M. Blouin1,2,3, Satish Kailasam Mani1,2,3, Christine Viaris de Lesegno1,2,3, Pierre Bost4,5, Alexandre Grassart6, Darius Köster7, Cesar Augusto Valades‑Cruz2,3,8, Valérie Chambon2,3,8, Ludger Johannes2,3,8, Paolo Pierobon9, Vassili Soumelis4, Catherine Coirault10, Stéphane Vassilopoulos10, and Christophe Lamaze1,2,3 Caveolae are small invaginated pits that function as dynamic mechanosensors to buffer tension variations at the plasma membrane. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Congenital Disorders of Glycosylation from a Neurological Perspective
brain sciences Review Congenital Disorders of Glycosylation from a Neurological Perspective Justyna Paprocka 1,* , Aleksandra Jezela-Stanek 2 , Anna Tylki-Szyma´nska 3 and Stephanie Grunewald 4 1 Department of Pediatric Neurology, Faculty of Medical Science in Katowice, Medical University of Silesia, 40-752 Katowice, Poland 2 Department of Genetics and Clinical Immunology, National Institute of Tuberculosis and Lung Diseases, 01-138 Warsaw, Poland; [email protected] 3 Department of Pediatrics, Nutrition and Metabolic Diseases, The Children’s Memorial Health Institute, W 04-730 Warsaw, Poland; [email protected] 4 NIHR Biomedical Research Center (BRC), Metabolic Unit, Great Ormond Street Hospital and Institute of Child Health, University College London, London SE1 9RT, UK; [email protected] * Correspondence: [email protected]; Tel.: +48-606-415-888 Abstract: Most plasma proteins, cell membrane proteins and other proteins are glycoproteins with sugar chains attached to the polypeptide-glycans. Glycosylation is the main element of the post- translational transformation of most human proteins. Since glycosylation processes are necessary for many different biological processes, patients present a diverse spectrum of phenotypes and severity of symptoms. The most frequently observed neurological symptoms in congenital disorders of glycosylation (CDG) are: epilepsy, intellectual disability, myopathies, neuropathies and stroke-like episodes. Epilepsy is seen in many CDG subtypes and particularly present in the case of mutations -

De Novo Mutations in Epileptic Encephalopathies
LETTER doi:10.1038/nature12439 De novo mutations in epileptic encephalopathies Epi4K Consortium* & Epilepsy Phenome/Genome Project* Epileptic encephalopathies are a devastating group of severe child- were found to be highly improbable (Table 1 and Fig. 1). We performed hood epilepsy disorders for which the cause is often unknown1.Here the same calculations on all of the genes with multiple de novo mutations we report a screen for de novo mutations in patients with two clas- observed in 610 control trios and found no genes with a significant excess sical epileptic encephalopathies: infantile spasms (n 5 149) and of de novo mutations (Supplementary Table 4). Although mutations in Lennox–Gastaut syndrome (n 5 115). We sequenced the exomes of GABRB3 have previously been reported in association with another type 264 probands, and their parents, and confirmed 329 de novo muta- of epilepsy15, and in vivo mouse studies suggest that GABRB3 haplo- tions. A likelihood analysis showed a significant excess of de novo insufficiency is one of the causes of epilepsy in Angelman’s syndrome16, mutations in the 4,000 genes that are the most intolerant to func- our observations implicate it, for the first time, as a single-gene cause of tional genetic variation in the human population (P 5 2.9 3 1023). epileptic encephalopathies and provide the strongest evidence to date for Among these are GABRB3,withde novo mutations in four patients, its association with any epilepsy. Likewise, ALG13, an X-linked gene and ALG13,withthesamede novo mutation in two patients; both encoding a subunit of the uridine diphosphate-N-acetylglucosamine genes show clear statistical evidence of association with epileptic transferase, was previously shown to carry a novel de novo mutation in encephalopathy. -

Supplemental Material For
Supplemental material for Epithelial to mesenchymal transition rewires the molecular path to PI3-Kinase-dependent proliferation Megan B. Salt1,2, Sourav Bandyopadhyay1,3 and Frank McCormick1 Authors Affiliations: 1-Helen Diller Family Comprehensive Cancer Center; 2-Biomedical Sciences Graduate Program, University of California San Francisco, San Francisco, California; 3-California Institute for Quantitative Biosciences, San Francisco, California. Supplemental Figure Legends Table S1. EMT associated changes in gene expression. Significantly (A) up- and (B) down- regulated genes in comparing H358-TwistER cells to control H358-GFP expressing cells treated with 100nM 4OHT for 12 days. The four columns on the right are associated with significantly upregulated genes, while the four columns furthest left described significantly downregulated genes. The first column for the up- and downregulated genes shows the Probe ID associated with each gene from the Affymetric Human Gene 1.0 ST microarrays. The gene name are shown in the next column, followed by the log2(fold change) in expression for each gene after 4OHT treatment in the H358-TwistER cells. The final column shows the p-value for each gene adjusted for the false discovery rate. Figure S1. Akt inhibition reduces serum-independent proliferation. (A) Proliferation of H358- TwistER cells with increasing concentrations (uM) of GSK-690693 (top) or MK-2206 (bottom). (B) Lysates from H358-TwistER cells treated for 48hrs in serum free media with indicated concentrations of GSK-690693 (top), or MK-2206 (bottom). Figure S2. The effects of TGFβ1 are specific to epithelial cells and lead to reduced serum- independent proliferation. (A) Proliferation of untreated (top) or TGFβ1 pre-treated (bottom) H358 and H441 cells in the presence (10%) or absence (SS) of serum. -

Supporting Information
Supporting Information Pouryahya et al. SI Text Table S1 presents genes with the highest absolute value of Ricci curvature. We expect these genes to have significant contribution to the network’s robustness. Notably, the top two genes are TP53 (tumor protein 53) and YWHAG gene. TP53, also known as p53, it is a well known tumor suppressor gene known as the "guardian of the genome“ given the essential role it plays in genetic stability and prevention of cancer formation (1, 2). Mutations in this gene play a role in all stages of malignant transformation including tumor initiation, promotion, aggressiveness, and metastasis (3). Mutations of this gene are present in more than 50% of human cancers, making it the most common genetic event in human cancer (4, 5). Namely, p53 mutations play roles in leukemia, breast cancer, CNS cancers, and lung cancers, among many others (6–9). The YWHAG gene encodes the 14-3-3 protein gamma, a member of the 14-3-3 family proteins which are involved in many biological processes including signal transduction regulation, cell cycle pro- gression, apoptosis, cell adhesion and migration (10, 11). Notably, increased expression of 14-3-3 family proteins, including protein gamma, have been observed in a number of human cancers including lung and colorectal cancers, among others, suggesting a potential role as tumor oncogenes (12, 13). Furthermore, there is evidence that loss Fig. S1. The histogram of scalar Ricci curvature of 8240 genes. Most of the genes have negative scalar Ricci curvature (75%). TP53 and YWHAG have notably low of p53 function may result in upregulation of 14-3-3γ in lung cancer Ricci curvatures. -

Associated with Low Dehydrodolichol Diphosphate Synthase (DHDDS) Activity S
Sabry et al. Orphanet Journal of Rare Diseases (2016) 11:84 DOI 10.1186/s13023-016-0468-1 RESEARCH Open Access A case of fatal Type I congenital disorders of glycosylation (CDG I) associated with low dehydrodolichol diphosphate synthase (DHDDS) activity S. Sabry1,2,3,4, S. Vuillaumier-Barrot1,2,5, E. Mintet1,2, M. Fasseu1,2, V. Valayannopoulos6, D. Héron7,8, N. Dorison8, C. Mignot7,8,9, N. Seta5,10, I. Chantret1,2, T. Dupré1,2,5 and S. E. H. Moore1,2* Abstract Background: Type I congenital disorders of glycosylation (CDG-I) are mostly complex multisystemic diseases associated with hypoglycosylated serum glycoproteins. A subgroup harbour mutations in genes necessary for the biosynthesis of the dolichol-linked oligosaccharide (DLO) precursor that is essential for protein N-glycosylation. Here, our objective was to identify the molecular origins of disease in such a CDG-Ix patient presenting with axial hypotonia, peripheral hypertonia, enlarged liver, micropenis, cryptorchidism and sensorineural deafness associated with hypo glycosylated serum glycoproteins. Results: Targeted sequencing of DNA revealed a splice site mutation in intron 5 and a non-sense mutation in exon 4 of the dehydrodolichol diphosphate synthase gene (DHDDS). Skin biopsy fibroblasts derived from the patient revealed ~20 % residual DHDDS mRNA, ~35 % residual DHDDS activity, reduced dolichol-phosphate, truncated DLO and N-glycans, and an increased ratio of [2-3H]mannose labeled glycoprotein to [2-3H]mannose labeled DLO. Predicted truncated DHDDS transcripts did not complement rer2-deficient yeast. SiRNA-mediated down-regulation of DHDDS in human hepatocellular carcinoma HepG2 cells largely mirrored the biochemical phenotype of cells from the patient. -

Evolutionary Fate of Retroposed Gene Copies in the Human Genome
Evolutionary fate of retroposed gene copies in the human genome Nicolas Vinckenbosch*, Isabelle Dupanloup*†, and Henrik Kaessmann*‡ *Center for Integrative Genomics, University of Lausanne, Ge´nopode, 1015 Lausanne, Switzerland; and †Computational and Molecular Population Genetics Laboratory, Zoological Institute, University of Bern, 3012 Bern, Switzerland Communicated by Wen-Hsiung Li, University of Chicago, Chicago, IL, December 30, 2005 (received for review December 14, 2005) Given that retroposed copies of genes are presumed to lack the and rodent genomes (7–12). In addition, three recent studies regulatory elements required for their expression, retroposition using EST data (13, 14) and tiling-microarray data from chro- has long been considered a mechanism without functional rele- mosome 22 (15) indicated that retrocopy transcription may be vance. However, through an in silico assay for transcriptional widespread, although these surveys were limited, and potential activity, we identify here >1,000 transcribed retrocopies in the functional implications were not addressed. human genome, of which at least Ϸ120 have evolved into bona To further explore the functional significance of retroposition fide genes. Among these, Ϸ50 retrogenes have evolved functions in the human genome, we systematically screened for signatures in testes, more than half of which were recruited as functional of selection related to retrocopy transcription. Our results autosomal counterparts of X-linked genes during spermatogene- suggest that retrocopy transcription is not rare and that the sis. Generally, retrogenes emerge ‘‘out of the testis,’’ because they pattern of transcription of human retrocopies has been pro- are often initially transcribed in testis and later evolve stronger and foundly shaped by natural selection, acting both for and against sometimes more diverse spatial expression patterns. -
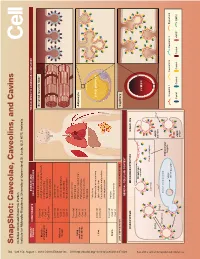
Snapshot: Caveolae, Caveolins, and Cavins Nicholas Ariotti and Robert G
704 Cell SnapShot: Caveolae, Caveolins, and Cavins Nicholas Ariotti and Robert G. Parton 154 Institute for Molecular Bioscience, University of Queensland, St. Lucia, QLD 4072, Australia , August 1,2013©2013 Elsevier Inc. DOI http://dx.doi.org/10.1016/j.cell.2013.07.009 ORGAN/ COMPONENTS DISEASE AND TISSUE-SPECIFIC CAVEOLAR COMPLEXES TISSUE CELLULAR PROCESS Rippling muscle disease Caveolin3 Striated muscle fiber Limb girdle muscular dystrophy Muscle Cavin1 skeletal and Cardiomyopathy Cavin4 cardiac Mechanoprotection Cav1 (heart) T-tubule formation Caveolin1 Lipodystrophy Caveolin2 Lipotoxicity Adipose Cavin1 Fatty acid regulation tissue Cavin2 Insulin signaling Cavin3 Mechanoprotection Atherosclerosis Caveolin1 Inammation Caveolin2 Lung Pulmonary hypertension and other Cavin1 Adipocyte Pulmonary brosis endothelia Cavin2 Mechanosensation Cavin3 Signaling Fatty liver Hepatocellular carcinoma Caveolin1 LIPID DROPLET Liver Lipid metabolism Caveolin2 Carbohydrate metabolism Liver regeneration Caveolin1 Autism Brain Cavin1 Schizophrenia Cavin3 LOW EXPRESSION HIGH EXPRESSION Capillary GENERAL CELLULAR CONTEXT ENDOCYTOSIS MECHANOPROTECTION SIGNALING See online version for legend and references. and legend for version online See Membrane stretch LUMEN Cavin complex Caveola EHD2 eNOS Ca2+ inactive Rab5 Dynamin2 EARLY ENDOSOME Intracellular targets MVB/ Caveolin 1 Caveolin 2 Caveolin 3 Dynamin2 late endosome eNOS Cavin1 Cavin2 Cavin3 Cavin4 eNOS EHD2 active SnapShot: Caveolae, Caveolins, and Cavins Nicholas Ariotti and Robert G. Parton Institute for Molecular Bioscience, University of Queensland, St. Lucia, QLD 4072, Australia Caveolae, submicroscopic bulb-shaped plasma membrane pits, are an abundant feature of many mammalian cells (Parton and del Pozo, 2013). Caveolae and the major proteins of caveolae, caveolins (Rothberg et al., 1992), and cavins (Hill et al., 2008), are linked to a number of human diseases such as muscular dystrophy, cardiomyopathy, and lipodys- trophy (see Glossary) (Bruno et al., 1993; Fernández et al., 2006; Hayashi et al., 2009). -

(Erf3/GSPT) in the Mrna Degradation Pathway
Biochemistry (Moscow), Vol. 64, No. 12, 1999, pp. 1367-1372. Translated from Biokhimiya, Vol. 64, No. 12, 1999, pp. 1621-1627. Original Russian Text Copyright © 1999 by Hoshino, Hosoda, Araki, Kobayashi, Uchida, Funakoshi, Katada. MINI-REVIEW Novel Function of the Eukaryotic Polypeptide-Chain Releasing Factor 3 (eRF3/GSPT) in the mRNA Degradation Pathway S. Hoshino*, N. Hosoda, Y. Araki, T. Kobayashi, N. Uchida, Y. Funakoshi, and T. Katada Department of Physiological Chemistry, Graduate School of Pharmaceutical Sciences, University of Tokyo, Tokyo 113-0033, Japan; fax: + 81 3-5841-4751; E-mail: [email protected] Received June 3, 1999 AbstractThe mammalian GTP-binding protein GSPT, whose carboxy-terminal sequence is homologous to the eukary- otic elongation factor EF1α, binds to the polypeptide chain releasing factor eRF1 to function as eRF3 in translation ter- mination. However, the amino-terminal domain of GSPT, which contains a prion-like sequence, is not required for the binding. Instead, the amino-terminal domain is capable of binding to the carboxy-terminal domain of polyadenylate- binding protein (PABP), whose amino terminus is associating with the poly(A) tail of mRNAs, presumably for their sta- bilization. Interestingly, multimerization of PABP with poly(A), which is ascribed to the action of its carboxy-terminal domain, was completely inhibited by the interaction with the amino-terminal domain of GSPT. This may facilitate short- ening of the poly(A) tail of mRNAs by an RNase. Thus, GSPT/eRF3 appears to function not only as a stimulator of eRF1 in the translation termination but also as an initiator of the mRNA degradation machinery.