Supplementary Information
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Reprogramming of Trna Modifications Controls the Oxidative Stress Response by Codon-Biased Translation of Proteins
Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation Chan, Clement T.Y. et al. “Reprogramming of tRNA Modifications Controls the Oxidative Stress Response by Codon-biased Translation of Proteins.” Nature Communications 3 (2012): 937. As Published http://dx.doi.org/10.1038/ncomms1938 Publisher Nature Publishing Group Version Author's final manuscript Citable link http://hdl.handle.net/1721.1/76775 Terms of Use Article is made available in accordance with the publisher's policy and may be subject to US copyright law. Please refer to the publisher's site for terms of use. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins Clement T.Y. Chan,1,2 Yan Ling Joy Pang,1 Wenjun Deng,1 I. Ramesh Babu,1 Madhu Dyavaiah,3 Thomas J. Begley3 and Peter C. Dedon1,4* 1Department of Biological Engineering, 2Department of Chemistry and 4Center for Environmental Health Sciences, Massachusetts Institute of Technology, Cambridge, MA 02139; 3College of Nanoscale Science and Engineering, University at Albany, SUNY, Albany, NY 12203 * Corresponding author: PCD, Department of Biological Engineering, NE47-277, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139; tel 617-253-8017; fax 617-324-7554; email [email protected] 2 ABSTRACT Selective translation of survival proteins is an important facet of cellular stress response. We recently demonstrated that this translational control involves a stress-specific reprogramming of modified ribonucleosides in tRNA. -

Allele-Specific Expression of Ribosomal Protein Genes in Interspecific Hybrid Catfish
Allele-specific Expression of Ribosomal Protein Genes in Interspecific Hybrid Catfish by Ailu Chen A dissertation submitted to the Graduate Faculty of Auburn University in partial fulfillment of the requirements for the Degree of Doctor of Philosophy Auburn, Alabama August 1, 2015 Keywords: catfish, interspecific hybrids, allele-specific expression, ribosomal protein Copyright 2015 by Ailu Chen Approved by Zhanjiang Liu, Chair, Professor, School of Fisheries, Aquaculture and Aquatic Sciences Nannan Liu, Professor, Entomology and Plant Pathology Eric Peatman, Associate Professor, School of Fisheries, Aquaculture and Aquatic Sciences Aaron M. Rashotte, Associate Professor, Biological Sciences Abstract Interspecific hybridization results in a vast reservoir of allelic variations, which may potentially contribute to phenotypical enhancement in the hybrids. Whether the allelic variations are related to the downstream phenotypic differences of interspecific hybrid is still an open question. The recently developed genome-wide allele-specific approaches that harness high- throughput sequencing technology allow direct quantification of allelic variations and gene expression patterns. In this work, I investigated allele-specific expression (ASE) pattern using RNA-Seq datasets generated from interspecific catfish hybrids. The objective of the study is to determine the ASE genes and pathways in which they are involved. Specifically, my study investigated ASE-SNPs, ASE-genes, parent-of-origins of ASE allele and how ASE would possibly contribute to heterosis. My data showed that ASE was operating in the interspecific catfish system. Of the 66,251 and 177,841 SNPs identified from the datasets of the liver and gill, 5,420 (8.2%) and 13,390 (7.5%) SNPs were identified as significant ASE-SNPs, respectively. -

Supplementary Materials
1 Supplementary Materials: Supplemental Figure 1. Gene expression profiles of kidneys in the Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice. (A) A heat map of microarray data show the genes that significantly changed up to 2 fold compared between Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice (N=4 mice per group; p<0.05). Data show in log2 (sample/wild-type). 2 Supplemental Figure 2. Sting signaling is essential for immuno-phenotypes of the Fcgr2b-/-lupus mice. (A-C) Flow cytometry analysis of splenocytes isolated from wild-type, Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice at the age of 6-7 months (N= 13-14 per group). Data shown in the percentage of (A) CD4+ ICOS+ cells, (B) B220+ I-Ab+ cells and (C) CD138+ cells. Data show as mean ± SEM (*p < 0.05, **p<0.01 and ***p<0.001). 3 Supplemental Figure 3. Phenotypes of Sting activated dendritic cells. (A) Representative of western blot analysis from immunoprecipitation with Sting of Fcgr2b-/- mice (N= 4). The band was shown in STING protein of activated BMDC with DMXAA at 0, 3 and 6 hr. and phosphorylation of STING at Ser357. (B) Mass spectra of phosphorylation of STING at Ser357 of activated BMDC from Fcgr2b-/- mice after stimulated with DMXAA for 3 hour and followed by immunoprecipitation with STING. (C) Sting-activated BMDC were co-cultured with LYN inhibitor PP2 and analyzed by flow cytometry, which showed the mean fluorescence intensity (MFI) of IAb expressing DC (N = 3 mice per group). 4 Supplemental Table 1. Lists of up and down of regulated proteins Accession No. -

Swissprot Genetrail Acc. No. ID Exp1 Exp2 Exp1 Exp2 Exp1 Exp2
Supplementary TABLE 1. Quantitative MS-based proteomic results from the macrophage cytosolic proteomes for both biological replicates separately Human proteins HSV-1 dLeu d3-4 Protein name SwissProt Genetrail N % Seq Cov Pept(95%) Fold change P-value Fold change P-value Fold change P-value Acc. No. ID Exp1 Exp2 Exp1 Exp2 Exp1 Exp2 Exp1 Exp2 Exp1 Exp2 Exp1 Exp2 Exp1 Exp2 Exp1 Exp2 Exp1 Exp2 10 kDa heat shock protein, mitochondrial P61604 HSPE1 121 108 69,6 65,7 9 18 1,24 1,11 0,014 0,141 1,12 1,16 0,127 0,054 1,07 0,96 0,290 0,529 14-3-3 protein beta/alpha P31946 YWHAB 350 145 37 52,9 13 16 1,10 0,97 0,409 0,743 1,12 0,72 0,651 0,184 1,04 0,72 0,730 0,101 14-3-3 protein epsilon P62258 YWHAE 661 524 16,5 24,7 5 8 1,43 1,00 0,996 0,60 0,78 0,238 0,84 0,87 0,765 14-3-3 protein eta Q04917 YWHAH 572 376 36,2 34,6 9 10 0,77 1,00 1,30 14-3-3 protein gamma P61981 YWHAG 135 358 38,1 35,2 13 10 1,93 1,21 0,170 1,10 0,44 0,515 1,88 1,17 0,099 14-3-3 protein theta P27348 YWHAQ 659 596 20,4 43,7 7 8 1,12 0,72 1,16 14-3-3 protein zeta/delta P63104 YWHAZ 59 36 45,3 54,7 15 19 1,48 1,33 0,004 0,019 1,22 0,95 0,265 0,570 1,08 1,10 0,315 0,416 182 kDa tankyrase-1-binding protein Q9C0C2 TNKS1BP1 517 2,7 2 0,89 0,874 1,18 0,345 1,06 0,651 1-phosphatidylinositol 4,5-bisphosphate phosphodiesteraseQ00722 beta-2 PLCB2 291 9,1 3 0,67 0,290 0,65 0,046 0,94 0,793 2,4-dienoyl-CoA reductase, mitochondrial Q16698 DECR1 335 14,3 4 0,76 0,284 0,97 0,841 0,91 0,415 26S proteasome non-ATPase regulatory subunit 11 O00231 PSMD11 375 18,7 3 1,43 1,19 0,93 26S proteasome non-ATPase -

Immunoprecipitation and Mass Spectrometry Defines an Extensive
BRES : 44759 Model7 pp: À 1221ðcol:fig: : NILÞ brain research ] ( ]]]]) ]]]– ]]] Available online at www.sciencedirect.com 121 122 123 124 125 126 www.elsevier.com/locate/brainres 127 128 129 Review 130 131 fi 132 Immunoprecipitation and mass spectrometry de nes 133 – 134 an extensive RBM45 protein protein interaction 135 Q2 136 network 137 138 a a,b a a c 139 Yang Li , Mahlon Collins , Jiyan An , Rachel Geiser , Tony Tegeler , c c c a,b,n 140 Q1 Kristine Tsantilas , Krystine Garcia , Patrick Pirrotte , Robert Bowser 141 aDivisions of Neurology and Neurobiology, Barrow Neurological Institute, St. Joseph's Hospital and Medical Center, 142 Phoenix, AZ 85013, USA 143 bUniversity of Pittsburgh School of Medicine, Pittsburgh, PA 15261, USA 144 cCenter for Proteomics, TGen (Translational Genomics Research Institute), Phoenix, AZ 85004, USA 145 146 147 article info abstract 148 149 Article history: The pathological accumulation of RNA-binding proteins (RBPs) within inclusion bodies is a 150 Received 30 January 2016 hallmark of amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration 151 Received in revised form (FTLD). RBP aggregation results in both toxic gain and loss of normal function. Determining 152 25 February 2016 the protein binding partners and normal functions of disease-associated RBPs is necessary 153 Accepted 28 February 2016 to fully understand molecular mechanisms of RBPs in disease. Herein, we characterized 154 the protein–protein interactions (PPIs) of RBM45, a RBP that localizes to inclusions in ALS/ 155 – fi Keywords: FTLD. Using immunoprecipitation coupled to mass spectrometry (IP MS), we identi ed 132 156 fi RBM45 proteins that speci cally interact with RBM45 within HEK293 cells. -
Download Validation Data
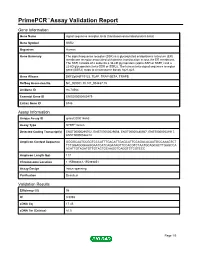
PrimePCR™Assay Validation Report Gene Information Gene Name signal sequence receptor, beta (translocon-associated protein beta) Gene Symbol SSR2 Organism Human Gene Summary The signal sequence receptor (SSR) is a glycosylated endoplasmic reticulum (ER) membrane receptor associated with protein translocation across the ER membrane. The SSR consists of 2 subunits a 34-kD glycoprotein (alpha-SSR or SSR1) and a 22-kD glycoprotein (beta-SSR or SSR2). The human beta-signal sequence receptor gene (SSR2) maps to chromosome bands 1q21-q23. Gene Aliases DKFZp686F19123, TLAP, TRAP-BETA, TRAPB RefSeq Accession No. NC_000001.10, NT_004487.19 UniGene ID Hs.74564 Ensembl Gene ID ENSG00000163479 Entrez Gene ID 6746 Assay Information Unique Assay ID qHsaCID0014663 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000295702, ENST00000529008, ENST00000480567, ENST00000531917, ENST00000526212 Amplicon Context Sequence GGGGCAATCCGGTCCCATTTGACATTGAGCATTCCAGACACAATGCCAAAGTCT TCTGGAGGGAAGGAATCATCAGATAGTTCCACGTCTAATGCAGCACTTGAGCCA ACATTGTAGATGTTGTACTGCAAGGTCAGGTCTCGTCCC Amplicon Length (bp) 117 Chromosome Location 1:155988061-155989851 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 98 R2 0.9998 cDNA Cq 17.45 cDNA Tm (Celsius) 81.5 Page 1/5 PrimePCR™Assay Validation Report gDNA Cq Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report SSR2, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal reference -

G Protein-Coupled Receptors
G PROTEIN-COUPLED RECEPTORS Overview:- The completion of the Human Genome Project allowed the identification of a large family of proteins with a common motif of seven groups of 20-24 hydrophobic amino acids arranged as α-helices. Approximately 800 of these seven transmembrane (7TM) receptors have been identified of which over 300 are non-olfactory receptors (see Frederikson et al., 2003; Lagerstrom and Schioth, 2008). Subdivision on the basis of sequence homology allows the definition of rhodopsin, secretin, adhesion, glutamate and Frizzled receptor families. NC-IUPHAR recognizes Classes A, B, and C, which equate to the rhodopsin, secretin, and glutamate receptor families. The nomenclature of 7TM receptors is commonly used interchangeably with G protein-coupled receptors (GPCR), although the former nomenclature recognises signalling of 7TM receptors through pathways not involving G proteins. For example, adiponectin and membrane progestin receptors have some sequence homology to 7TM receptors but signal independently of G-proteins and appear to reside in membranes in an inverted fashion compared to conventional GPCR. Additionally, the NPR-C natriuretic peptide receptor has a single transmembrane domain structure, but appears to couple to G proteins to generate cellular responses. The 300+ non-olfactory GPCR are the targets for the majority of drugs in clinical usage (Overington et al., 2006), although only a minority of these receptors are exploited therapeutically. Signalling through GPCR is enacted by the activation of heterotrimeric GTP-binding proteins (G proteins), made up of α, β and γ subunits, where the α and βγ subunits are responsible for signalling. The α subunit (tabulated below) allows definition of one series of signalling cascades and allows grouping of GPCRs to suggest common cellular, tissue and behavioural responses. -

Aneuploidy: Using Genetic Instability to Preserve a Haploid Genome?
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Science (Cancer Biology) Aneuploidy: Using genetic instability to preserve a haploid genome? Submitted by: Ramona Ramdath In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Science Examination Committee Signature/Date Major Advisor: David Allison, M.D., Ph.D. Academic James Trempe, Ph.D. Advisory Committee: David Giovanucci, Ph.D. Randall Ruch, Ph.D. Ronald Mellgren, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: April 10, 2009 Aneuploidy: Using genetic instability to preserve a haploid genome? Ramona Ramdath University of Toledo, Health Science Campus 2009 Dedication I dedicate this dissertation to my grandfather who died of lung cancer two years ago, but who always instilled in us the value and importance of education. And to my mom and sister, both of whom have been pillars of support and stimulating conversations. To my sister, Rehanna, especially- I hope this inspires you to achieve all that you want to in life, academically and otherwise. ii Acknowledgements As we go through these academic journeys, there are so many along the way that make an impact not only on our work, but on our lives as well, and I would like to say a heartfelt thank you to all of those people: My Committee members- Dr. James Trempe, Dr. David Giovanucchi, Dr. Ronald Mellgren and Dr. Randall Ruch for their guidance, suggestions, support and confidence in me. My major advisor- Dr. David Allison, for his constructive criticism and positive reinforcement. -

A Crosstalk Between the RNA Binding Protein Smaug and the Hedgehog Pathway Links Cell Signaling to Mrna Regulation in Drosophila Lucía Bruzzone
A crosstalk between the RNA binding protein Smaug and the Hedgehog pathway links cell signaling to mRNA regulation in drosophila Lucía Bruzzone To cite this version: Lucía Bruzzone. A crosstalk between the RNA binding protein Smaug and the Hedgehog pathway links cell signaling to mRNA regulation in drosophila. Cellular Biology. Université Sorbonne Paris Cité, 2018. English. NNT : 2018USPCC234. tel-02899776 HAL Id: tel-02899776 https://tel.archives-ouvertes.fr/tel-02899776 Submitted on 15 Jul 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Thèse de doctorat de l’Université Sorbonne Paris Cité Préparée à l’Université Paris Diderot Ecole doctorale HOB n° 561 Institut Jacques Monod / Equipe Développement, Signalisation et Trafic A crosstalk between the RNA binding protein Smaug and the Hedgehog pathway links cell signaling to mRNA regulation in Drosophila Lucía Bruzzone Thèse de doctorat de Biologie Dirigée par Anne Plessis Présentée et soutenue publiquement à Paris le 19 mars 2018 Président du jury: Alain Zider / Professeur Université Paris Diderot -

Anti-GNB4 (GW21993)
3050 Spruce Street, Saint Louis, MO 63103 USA Tel: (800) 521-8956 (314) 771-5765 Fax: (800) 325-5052 (314) 771-5757 email: [email protected] Product Information Anti-GNB4 antibody produced in chicken, affinity isolated antibody Catalog Number GW21993 Formerly listed as GenWay Catalog Number 15-288-21993, Guanine nucleotide-binding protein subunit beta 4 Anti- body. The product is a clear, colorless solution in phosphate – Storage Temperature Store at 20 °C buffered saline, pH 7.2, containing 0.02% sodium azide. Synonyms: Guanine nucleotide-binding protein, beta-4 Species Reactivity: Human, mouse subunit, Transducin beta chain 4 Tested Applications: WB Product Description Recommended Dilutions: Recommended starting dilution Guanine nucleotide-binding proteins (G proteins) are for Western blot analysis is 1:500, for tissue or cell staining involved as a modulator or transducer in various trans- 1:200. membrane signaling systems. The beta and gamma chains are required for the GTPase activity. for replacement of GDP Note: Optimal concentrations and conditions for each by GTP. and for G protein-effector interaction. application should be determined by the user. NCBI Accession number: NP_067642.1 Precautions and Disclaimer Swiss Prot Accession number: Q9HAV0 This product is for R&D use only, not for drug, household, or other uses. Due to the sodium azide content a material Gene Information: Human .. GNB4 (59345) safety data sheet (MSDS) for this product has been sent to Immunogen: Recombinant protein Guanine nucleotide- the attention of the safety officer of your institution. Please binding protein, beta-4 subunit consult the Material Safety Data Sheet for information regarding hazards and safe handling practices. -

Disease-Related Cellular Protein Networks Differentially Affected
www.nature.com/scientificreports OPEN Disease‑related cellular protein networks diferentially afected under diferent EGFR mutations in lung adenocarcinoma Toshihide Nishimura1,8*, Haruhiko Nakamura1,2,8, Ayako Yachie3,8, Takeshi Hase3,8, Kiyonaga Fujii1,8, Hirotaka Koizumi4, Saeko Naruki4, Masayuki Takagi4, Yukiko Matsuoka3, Naoki Furuya5, Harubumi Kato6,7 & Hisashi Saji2 It is unclear how epidermal growth factor receptor EGFR major driver mutations (L858R or Ex19del) afect downstream molecular networks and pathways. This study aimed to provide information on the infuences of these mutations. The study assessed 36 protein expression profles of lung adenocarcinoma (Ex19del, nine; L858R, nine; no Ex19del/L858R, 18). Weighted gene co-expression network analysis together with analysis of variance-based screening identifed 13 co-expressed modules and their eigen proteins. Pathway enrichment analysis for the Ex19del mutation demonstrated involvement of SUMOylation, epithelial and mesenchymal transition, ERK/mitogen- activated protein kinase signalling via phosphorylation and Hippo signalling. Additionally, analysis for the L858R mutation identifed various pathways related to cancer cell survival and death. With regard to the Ex19del mutation, ROCK, RPS6KA1, ARF1, IL2RA and several ErbB pathways were upregulated, whereas AURK and GSKIP were downregulated. With regard to the L858R mutation, RB1, TSC22D3 and DOCK1 were downregulated, whereas various networks, including VEGFA, were moderately upregulated. In all mutation types, CD80/CD86 (B7), MHC, CIITA and IFGN were activated, whereas CD37 and SAFB were inhibited. Costimulatory immune-checkpoint pathways by B7/CD28 were mainly activated, whereas those by PD-1/PD-L1 were inhibited. Our fndings may help identify potential therapeutic targets and develop therapeutic strategies to improve patient outcomes. -
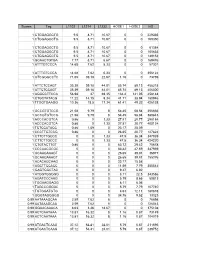
Scores Tag L1102 L1214 L1232 HOSE1 HOSE2 HS 1
Scores Tag L1102 L1214 L1232 HOSE1 HOSE2 HS 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 229335 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 169350 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 61384 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 105633 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 149152 1 GCAACTGTGA 7.77 8.71 6.67 0 0 169476 1 ATTTGTCCCA 14.68 7.62 5.33 0 0 57301 1 ATTTGTCCCA 14.68 7.62 5.33 0 0 356122 1 GTCGGGCCTC 71.65 39.18 22.67 1.16 0 73769 1 ATTCTCCAGT 35.39 39.18 44.01 85.74 89.13 458218 1 ATTCTCCAGT 35.39 39.18 44.01 85.74 89.13 406300 1 AGGGCTTCCA 56.98 37 69.35 134.4 141.05 458148 1 CTGCTATACG 11.22 14.15 9.34 41.71 38.94 180946 1 TTGGTGAAGG 10.36 18.5 17.34 61.41 49.32 426138 1 GCCGTGTCCG 21.58 9.79 8 54.45 58.84 356666 1 GCCGTGTCCG 21.58 9.79 8 54.45 58.84 380843 1 ACCCACGTCA 0.86 0 1.33 27.81 20.77 298184 1 ACCCACGTCA 0.86 0 1.33 27.81 20.77 400124 1 TCTCCATACC 0.86 1.09 0 23.17 25.09 1 CCCTTGTCCG 0.86 0 0 26.65 20.77 127824 1 CTTCTTGCCC 0 0 1.33 47.5 36.34 347939 1 CTTCTTGCCC 0 0 1.33 47.5 36.34 424220 1 CTGTACTTGT 0.86 0 0 63.72 29.42 75678 1 CCCAACGCGC 0 0 0 83.42 47.59 347939 1 GCAAGAAAGT 0 0 0 26.65 39.81 36977 1 GCAAGAAAGT 0 0 0 26.65 39.81 155376 1 ACACAGCAAG 0 0 0 23.17 15.58 1 AGCTTCCACC 0 0 0 11.59 7.79 355542 1 GAGTGGCTAC 0 0 0 9.27 6.92 1 ATGGTGGGGG 0 0 0 8.11 22.5 343586 1 AGATCCCAAG 0 0 0 5.79 8.65 50813 1 TGGAAGGAGG 0 0 0 8.11 6.06 1 TAGCCGGGAC 0 0 0 5.79 7.79 107740 1 TGTGGATGTG 0 0 0 4.63 12.11 180878 1 GGGTAGGGGG 0 0 0 34.76 9.52 13323 0.99 AATAAAGCAA 2.59 7.62 8 0 0 76698 0.99 AATAAAGCAA 2.59 7.62 8 0 0 126043 0.99 GGAACAAACA 8.63 3.26 18.67 0 0 375108