PKBΑ Isoform Signaling
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Profiling Data
Compound Name DiscoveRx Gene Symbol Entrez Gene Percent Compound Symbol Control Concentration (nM) JNK-IN-8 AAK1 AAK1 69 1000 JNK-IN-8 ABL1(E255K)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317I)-nonphosphorylated ABL1 87 1000 JNK-IN-8 ABL1(F317I)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317L)-nonphosphorylated ABL1 65 1000 JNK-IN-8 ABL1(F317L)-phosphorylated ABL1 61 1000 JNK-IN-8 ABL1(H396P)-nonphosphorylated ABL1 42 1000 JNK-IN-8 ABL1(H396P)-phosphorylated ABL1 60 1000 JNK-IN-8 ABL1(M351T)-phosphorylated ABL1 81 1000 JNK-IN-8 ABL1(Q252H)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(Q252H)-phosphorylated ABL1 56 1000 JNK-IN-8 ABL1(T315I)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(T315I)-phosphorylated ABL1 92 1000 JNK-IN-8 ABL1(Y253F)-phosphorylated ABL1 71 1000 JNK-IN-8 ABL1-nonphosphorylated ABL1 97 1000 JNK-IN-8 ABL1-phosphorylated ABL1 100 1000 JNK-IN-8 ABL2 ABL2 97 1000 JNK-IN-8 ACVR1 ACVR1 100 1000 JNK-IN-8 ACVR1B ACVR1B 88 1000 JNK-IN-8 ACVR2A ACVR2A 100 1000 JNK-IN-8 ACVR2B ACVR2B 100 1000 JNK-IN-8 ACVRL1 ACVRL1 96 1000 JNK-IN-8 ADCK3 CABC1 100 1000 JNK-IN-8 ADCK4 ADCK4 93 1000 JNK-IN-8 AKT1 AKT1 100 1000 JNK-IN-8 AKT2 AKT2 100 1000 JNK-IN-8 AKT3 AKT3 100 1000 JNK-IN-8 ALK ALK 85 1000 JNK-IN-8 AMPK-alpha1 PRKAA1 100 1000 JNK-IN-8 AMPK-alpha2 PRKAA2 84 1000 JNK-IN-8 ANKK1 ANKK1 75 1000 JNK-IN-8 ARK5 NUAK1 100 1000 JNK-IN-8 ASK1 MAP3K5 100 1000 JNK-IN-8 ASK2 MAP3K6 93 1000 JNK-IN-8 AURKA AURKA 100 1000 JNK-IN-8 AURKA AURKA 84 1000 JNK-IN-8 AURKB AURKB 83 1000 JNK-IN-8 AURKB AURKB 96 1000 JNK-IN-8 AURKC AURKC 95 1000 JNK-IN-8 -

Coordinate Phosphorylation of Multiple Residues on Single AKT1 and AKT2 Molecules
Oncogene (2014) 33, 3463–3472 & 2014 Macmillan Publishers Limited All rights reserved 0950-9232/14 www.nature.com/onc ORIGINAL ARTICLE Coordinate phosphorylation of multiple residues on single AKT1 and AKT2 molecules H Guo1,6, M Gao1,6,YLu1, J Liang1, PL Lorenzi2, S Bai3, DH Hawke4,JLi1, T Dogruluk5, KL Scott5, E Jonasch3, GB Mills1 and Z Ding1 Aberrant AKT activation is prevalent across multiple human cancer lineages providing an important new target for therapy. Twenty- two independent phosphorylation sites have been identified on specific AKT isoforms likely contributing to differential isoform regulation. However, the mechanisms regulating phosphorylation of individual AKT isoform molecules have not been elucidated because of the lack of robust approaches able to assess phosphorylation of multiple sites on a single AKT molecule. Using a nanofluidic proteomic immunoassay (NIA), consisting of isoelectric focusing followed by sensitive chemiluminescence detection, we demonstrate that under basal and ligand-induced conditions that the pattern of phosphorylation events is markedly different between AKT1 and AKT2. Indeed, there are at least 12 AKT1 peaks and at least 5 AKT2 peaks consistent with complex combinations of phosphorylation of different sites on individual AKT molecules. Following insulin stimulation, AKT1 was phosphorylated at Thr308 in the T-loop and Ser473 in the hydrophobic domain. In contrast, AKT2 was only phosphorylated at the equivalent sites (Thr309 and Ser474) at low levels. Further, Thr308 and Ser473 phosphorylation occurred predominantly on the same AKT1 molecules, whereas Thr309 and Ser474 were phosphorylated primarily on different AKT2 molecules. Although basal AKT2 phosphorylation was sensitive to inhibition of phosphatidylinositol 3-kinase (PI3K), basal AKT1 phosphorylation was essentially resistant. -

Divergent Regulation of Akt1 and Akt2 Isoforms in Insulin Target Tissues of Obese Zucker Rats Young-Bum Kim, Odile D
Divergent Regulation of Akt1 and Akt2 Isoforms in Insulin Target Tissues of Obese Zucker Rats Young-Bum Kim, Odile D. Peroni, Thomas F. Franke, and Barbara B. Kahn To determine whether impaired Akt (protein kinase B or rac) activation contributes to insulin resistance in vivo, we examined the expression, phosphorylation, he effect of insulin to acutely stimulate glucose and kinase activities of Akt1 and Akt2 isoforms in uptake and metabolism in peripheral tissues is insulin target tissues of insulin-resistant obese Zucker essential for normal glucose homeostasis. Resis- ؋ rats. In lean rats, insulin (10 U/kg i.v. 2.5 min) stim- tance to this effect is a major pathogenic feature ulated Akt1 activity 6.2-, 8.8-, and 4.4-fold and Akt2 T of type 2 diabetes (1,2) and contributes to the morbidity of activity 5.4-, 9.3-, and 1.8-fold in muscle, liver, and adi- pose tissue, respectively. In obese rats, insulin-stimu- obesity as well as type 1 diabetes (3). Whereas many of the lated Akt1 activity decreased 30% in muscle and 21% proximal steps in insulin signaling have been identified, the in adipose tissue but increased 37% in liver compared downstream pathways for insulin action to maintain glucose with lean littermates. Insulin-stimulated Akt2 activity homeostasis are still unknown. Insulin action involves a decreased 29% in muscle and 37% in liver but series of signaling cascades initiated by insulin binding to its increased 24% in adipose tissue. Akt2 protein levels receptor and eliciting receptor autophosphorylation and acti- were reduced 56% in muscle and 35% in liver of obese vation of receptor tyrosine kinases, which result in tyrosine rats, but Akt1 expression was unaltered. -

In Vitro and in Vivo Profile of a Selective, Allosteric Akt2 Inhibitor
In Vitro and In Vivo Profile of a Selective, Allosteric Akt2 Inhibitor Martin O'Rourke1, Caroline Boyd1, Estelle McLean1, Natalie Page1, Shane Rountree1, Steven Shepherd1, Mary McFarland1 Colin O’Dowd1, Graham Trevitt1, Iain James1, Timothy Harrison1,2, Ultan McDermott3 1Almac Discovery, Craigavon, United Kingdom; 2Centre for Cancer Research and Cell Biology, Queens University, Belfast, United Kingdom 3 Wellcome Trust Sanger Institute, Hinxton, Cambs, CB10 1SA Biochemical efficacy of ADC-0008830, an Akt2 selective Pathway biomarker Overview inhibitor engagement in vitro Figure 1: Dose dependent inhibition of a) Akt1 b) Akt2 and c) Akt3 in an FP Figure 2: Inhibition of pathway • Akt protein family members (Akt biochemical assay biomarkers by ELISA 1-3) are involved in a wide variety a b c pGSK3β pPRAS 40 of biological processes including Cell line pAkt (nM) cell proliferation, apoptosis, and (nM) (nM) tumorigenesis. PC3 60 100 15 (IC50 7,600nM) (IC 4,200nM) (IC 27nM) • We have identified a novel, 50 50 LnCaP 500 6000 3600 allosteric Akt 2 selective inhibitor, A2780 160 740 840 which exhibits single agent A549 130 129 150 efficacy in a panel of cell-lines. Panc-1 9 234 Not tested • In collaboration with the Sanger • ADC-0008830 is a potent inhibitor of the Akt2 isoform Institute, we have identified genes • ADC-0008830 inhibits key • No activity observed against PH domain null Akt isoforms confirming of sensitivity and resistance for components of the Akt pathway an allosteric mode of action subtype selective Akt inhibitors in multiple cell lines • Excellent selectivity against a kinase counterscreen panel (data not shown) Dose and time dependent inhibition of Cellular profiling of ADC-0008939, a prototypical isoform selective Akt2 pathway biomarkers (Panc-1) inhibitor, in the Sanger cell line panel Figure 3: Dose and time dependent inhibition of Akt Figure 4: Pharmacogenomic analysis in 432 cell lines of ADC-0008939, a prototypical isoform selective pathway in Panc-1 cell line Akt inhibitor (IC50 Akt1 300nM; Akt2 7nM; Akt3 1230nM). -

Supplemental Table 1: Snps Genotyped for NCO, Listed Alphabetically by Gene Name
Supplemental Table 1: SNPs genotyped for NCO, listed alphabetically by gene name. Gene Name SNP rs# ACVR1 rs10497189 ACVR1 rs10497191 ACVR1 rs10497192 ACVR1 rs10933441 ACVR1 rs1146035 ACVR1 rs1220110 ACVR1 rs17182166 ACVR1 rs17798043 ACVR1 rs4380178 ACVR1 rs6719924 ACVR2 rs1128919 ACVR2 rs10497025 ACVR2 rs1424941 ACVR2 rs1424954 ACVR2 rs17742573 ACVR2 rs2382112 ACVR2 rs4419186 AKT2 rs11671439 AKT2 rs12460555 AKT2 rs16974157 AKT2 rs2304188 AKT2 rs3730050 AKT2 rs7250897 AKT2 rs7254617 AKT2 rs874269 ALOX12B/ALOXE3 rs3809881 ALOX15B rs4792147 ALOX15B rs9898751 AMH rs10407022 AMH rs2074860 AMH rs3746158 AMH rs4806834 AMH rs757595 AMH rs886363 AMHR2 rs10876451 AMHR2 rs11170547 AMHR2 rs11170558 APC rs2229992 APC rs351771 APC rs41115 APC rs42427 APC rs459552 APC rs465899 APEX1 rs2275007 APEX1 rs3136820 15 APEX1 rs938883 APEX1 rs11160711 APEX1 rs1713459 APEX1 rs1713460 APEX1 rs1760941 APEX1 rs2275008 APEX1 rs3120073 APEX1 rs4465523 AR rs12011793 AR rs1204038 AR rs1337080 AR rs1337082 AR rs2207040 AR rs2361634 AR rs5031002 AR rs6152 AR rs962458 ATM rs1800058 ATM rs1800889 ATM rs11212570 ATM rs17503908 ATM rs227060 ATM rs228606 ATM rs3092991 ATM rs4987876 ATM rs4987886 ATM rs4987923 ATM rs4988023 ATM rs611646 ATM rs639923 ATM rs672655 ATR rs2229032 ATR rs10804682 ATR rs11920625 ATR rs13065800 ATR rs13085998 ATR rs13091637 ATR rs1802904 ATR rs6805118 ATR rs9856772 BACH1 rs388707 BACH1 rs1153276 BACH1 rs1153280 BACH1 rs1153284 BACH1 rs1153285 BACH1 rs17743655 BACH1 rs17744121 BACH1 rs2300301 BACH1 rs2832283 16 BACH1 rs411697 BACH1 rs425989 BARD1 rs1048108 -

Activation of Diverse Signalling Pathways by Oncogenic PIK3CA Mutations
ARTICLE Received 14 Feb 2014 | Accepted 12 Aug 2014 | Published 23 Sep 2014 DOI: 10.1038/ncomms5961 Activation of diverse signalling pathways by oncogenic PIK3CA mutations Xinyan Wu1, Santosh Renuse2,3, Nandini A. Sahasrabuddhe2,4, Muhammad Saddiq Zahari1, Raghothama Chaerkady1, Min-Sik Kim1, Raja S. Nirujogi2, Morassa Mohseni1, Praveen Kumar2,4, Rajesh Raju2, Jun Zhong1, Jian Yang5, Johnathan Neiswinger6, Jun-Seop Jeong6, Robert Newman6, Maureen A. Powers7, Babu Lal Somani2, Edward Gabrielson8, Saraswati Sukumar9, Vered Stearns9, Jiang Qian10, Heng Zhu6, Bert Vogelstein5, Ben Ho Park9 & Akhilesh Pandey1,8,9 The PIK3CA gene is frequently mutated in human cancers. Here we carry out a SILAC-based quantitative phosphoproteomic analysis using isogenic knockin cell lines containing ‘driver’ oncogenic mutations of PIK3CA to dissect the signalling mechanisms responsible for oncogenic phenotypes induced by mutant PIK3CA. From 8,075 unique phosphopeptides identified, we observe that aberrant activation of PI3K pathway leads to increased phosphorylation of a surprisingly wide variety of kinases and downstream signalling networks. Here, by integrating phosphoproteomic data with human protein microarray-based AKT1 kinase assays, we discover and validate six novel AKT1 substrates, including cortactin. Through mutagenesis studies, we demonstrate that phosphorylation of cortactin by AKT1 is important for mutant PI3K-enhanced cell migration and invasion. Our study describes a quantitative and global approach for identifying mutation-specific signalling events and for discovering novel signalling molecules as readouts of pathway activation or potential therapeutic targets. 1 McKusick-Nathans Institute of Genetic Medicine and Department of Biological Chemistry, Johns Hopkins University School of Medicine, 733 North Broadway, BRB 527, Baltimore, Maryland 21205, USA. -

Inhibitors of Cyclin-Dependent Kinases: Types and Their Mechanism of Action
International Journal of Molecular Sciences Review Inhibitors of Cyclin-Dependent Kinases: Types and Their Mechanism of Action Paweł Łukasik 1, Irena Baranowska-Bosiacka 2, Katarzyna Kulczycka 3 and Izabela Gutowska 1,* 1 Department of Medical Chemistry, Pomeranian Medical University in Szczecin, Powstancow Wlkp. 72 Av., 70-111 Szczecin, Poland; [email protected] 2 Department of Biochemistry, Pomeranian Medical University in Szczecin, Powstancow Wlkp. 72 Av., 70-111 Szczecin, Poland; [email protected] 3 Department of Pharmaceutical Chemistry, Pomeranian Medical University in Szczecin, Powstancow Wlkp. 72 Av., 70-111 Szczecin, Poland; [email protected] * Correspondence: [email protected] Abstract: Recent studies on cyclin-dependent kinase (CDK) inhibitors have revealed that small molecule drugs have become very attractive for the treatment of cancer and neurodegenerative disorders. Most CDK inhibitors have been developed to target the ATP binding pocket. However, CDK kinases possess a very similar catalytic domain and three-dimensional structure. These features make it difficult to achieve required selectivity. Therefore, inhibitors which bind outside the ATP binding site present a great interest in the biomedical field, both from the fundamental point of view and for the wide range of their potential applications. This review tries to explain whether the ATP competitive inhibitors are still an option for future research, and highlights alternative approaches to discover more selective and potent small molecule inhibitors. Keywords: cyclin-dependent kinase inhibitors; cancer; cell cycle; CDKs; CDK inhibitors Citation: Łukasik, P.; Baranowska-Bosiacka, I.; Kulczycka, K.; Gutowska, I. Inhibitors of Cyclin-Dependent Kinases: Types 1. Cyclin-Dependent Kinases (CDKs) and Their Mechanism of Action. -

PIK3CA and AKT2 Mutations of Gastric Cancer in China
PIK3CA and AKT2 mutations of gastric cancer in China WEN-XIANG CHENG1, 2#, Taimei Zhou1, 3#, FU-HUA PI1, YONG MENG4, WILLIAM AU1, QING-YING ZHANG1* # These authors contributed equally to this work. 1. Department of Preventive Medicine, Shantou University Medical College, Shantou 515041, China. 2. Translational Medicine R&D Center, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, 518055, China 3. Department of Laboratory Medicine, Huaihua Medical College, Huaihua, Hunan 418000, China 4. Department of Surgery, the First Affiliated Hospital of Shantou University Medical College, Shantou, Guangdong 515041, China. chinaXiv:201803.00958v1 * Correspondence to: Dr Qingying Zhang, Department of Preventive Medicine, Shantou University Medical College, 22# Xinling Road , Shantou 515041, P.R.China. Tel: (86754) 8890-0445 Fax: (86754) 8855-7562 E-mail address: [email protected] - 1 - Abstract Mutations in PI3K and/or AKT have been reported in a variety of cancers. This indicates that the two pathways interact to cause cancer. We have therefore investigated their roles in gastric cancer (GC) in China. In our study, exons 9, 18 and 20 of PIK3CA gene and exons 6~14 of AKT2 gene were screened in 10 GC cell lines and 100 advanced primary GC together with matched normal tissues. Denaturing high performance liquid chromatography (DHPLC) and DNA sequencing were used to analyze the mutations in the two genes. Two point mutations in the PIK3CA gene were identified in 4 of 10 GC cell lines and in 4 of 100 GC primary tumors. Two polymorphisms in AKT2 were detected in 19 of 100 GC primary tumors. -

PRODUCTS and SERVICES Target List
PRODUCTS AND SERVICES Target list Kinase Products P.1-11 Kinase Products Biochemical Assays P.12 "QuickScout Screening Assist™ Kits" Kinase Protein Assay Kits P.13 "QuickScout Custom Profiling & Panel Profiling Series" Targets P.14 "QuickScout Custom Profiling Series" Preincubation Targets Cell-Based Assays P.15 NanoBRET™ TE Intracellular Kinase Cell-Based Assay Service Targets P.16 Tyrosine Kinase Ba/F3 Cell-Based Assay Service Targets P.17 Kinase HEK293 Cell-Based Assay Service ~ClariCELL™ ~ Targets P.18 Detection of Protein-Protein Interactions ~ProbeX™~ Stable Cell Lines Crystallization Services P.19 FastLane™ Structures ~Premium~ P.20-21 FastLane™ Structures ~Standard~ Kinase Products For details of products, please see "PRODUCTS AND SERVICES" on page 1~3. Tyrosine Kinases Note: Please contact us for availability or further information. Information may be changed without notice. Expression Protein Kinase Tag Carna Product Name Catalog No. Construct Sequence Accession Number Tag Location System HIS ABL(ABL1) 08-001 Full-length 2-1130 NP_005148.2 N-terminal His Insect (sf21) ABL(ABL1) BTN BTN-ABL(ABL1) 08-401-20N Full-length 2-1130 NP_005148.2 N-terminal DYKDDDDK Insect (sf21) ABL(ABL1) [E255K] HIS ABL(ABL1)[E255K] 08-094 Full-length 2-1130 NP_005148.2 N-terminal His Insect (sf21) HIS ABL(ABL1)[T315I] 08-093 Full-length 2-1130 NP_005148.2 N-terminal His Insect (sf21) ABL(ABL1) [T315I] BTN BTN-ABL(ABL1)[T315I] 08-493-20N Full-length 2-1130 NP_005148.2 N-terminal DYKDDDDK Insect (sf21) ACK(TNK2) GST ACK(TNK2) 08-196 Catalytic domain -
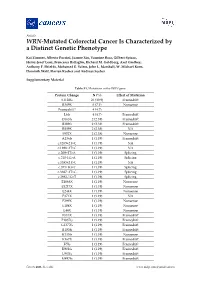
WRN-Mutated Colorectal Cancer Is Characterized by a Distinct Genetic Phenotype
Article WRN-Mutated Colorectal Cancer Is Characterized by a Distinct Genetic Phenotype Kai Zimmer, Alberto Puccini, Joanne Xiu, Yasmine Baca, Gilbert Spizzo, Heinz-Josef Lenz, Francesca Battaglin, Richard M. Goldberg, Axel Grothey, Anthony F. Shields, Mohamed E. Salem, John L. Marshall, W. Michael Korn, Dominik Wolf, Florian Kocher and Andreas Seeber Supplementary Material Table S1. Mutations in the WRN gene. Protein Change N (%) Effect of Mutation S1128fs 26 (30.9) Frameshift R369X 6 (7.1) Nonsense Frameshift* 4 (4.7) L6fs 4 (4.7) Frameshift D163fs 2 (2.38) Frameshift R389fs 2 (2.38) Frameshift R889X 2 (2.38) NA S952X 2 (2.38) Nonsense A136fs 1 (1.19) Frameshift c.1269+2T>C 1 (1.19) NA c.1898+2T>G 1 (1.19) NA c.209+2T>A 1 (1.19) Splicing c.210-1G>A 1 (1.19) Splicing c.3383+2T>C 1 (1.19) NA c.355+1G>T 1 (1.19) Splicing c.3687+2T>G 1 (1.19) Splicing c.3983-1G>T 1 (1.19) Splicing E1068X 1 (1.19) Nonsense E1217X 1 (1.19) Nonsense E244X 1 (1.19) Nonsense E371X 1 (1.19) NA E399X 1 (1.19) Nonsense E488X 1 (1.19) Nonsense E48X 1 (1.19) Nonsense E513X 1 (1.19) Frameshift F1037fs 1 (1.19) Frameshift G1377fs 1 (1.19) Frameshift I1183fs 1 (1.19) Frameshift K135fs 1 (1.19) Nonsense K167X 1 (1.19) Frameshift K5fs 1 (1.19) Frameshift K901fs 1 (1.19) Frameshift L967fs 1 (1.19) Frameshift M497fs 1 (1.19) Frameshift Cancers 2020, 12, x; doi: www.mdpi.com/journal/cancers Cancers 2020, 12 S2 of 13 N166fs 1 (1.19) Frameshift Q11fs 1 (1.19) Frameshift R1305X 1 (1.19) Nonsense R279fs 1 (1.19) Frameshift R565X 1 (1.19) Nonsene R741X 1 (1.19) Nonsense R987X 1 (1.19) NA T1011fs 1 (1.19) Frameshift W1014X 1 (1.19) Nonsense Y57X 1 (1.19) Nonsense Grand Total 84 *4 Frameshift mutations could not be described more precisely. -
Oncomine Comprehensive Assay V3 Empower Your Oncology Research with Proven Ion Torrent Technology
Oncomine Comprehensive Assay v3 Empower your oncology research with proven Ion Torrent technology The Oncomine Comprehensive Assay v3 is available for use on the Ion GeneStudio S5 Systems with Ion Chef Instrument or for use on the Genexus System The Ion Torrent™ Oncomine™ Comprehensive Assay v3 • Detects relevant SNVs, CNVs, gene fusions, and is a member of the family of Ion Torrent™ Oncomine™ indels from 161 unique cancer driver genes in one assays for clinical cancer research. Oncomine assays streamlined workfl ow are multiple-biomarker assays based on next-generation • Optimized and verifi ed for the Ion Chef™ Instrument and sequencing (NGS). They have been adopted by leading Ion GeneStudio™ S5 Systems with the Ion 540™ Chip, cancer institutions around the world, have been used to or the new Ion Torrent™ Genexus™ System with the profi le thousands of samples in diff erent translational and Ion Torrent™ GX5™ Chip, both enabling full automation clinical research projects, and have consistently delivered including automated library prep reliable results. “The requirement of a lower DNA input for the Oncomine assay Oncomine Comprehensive Assay v3 is a signifi cant advantage when primary samples are becoming • Content is based on the latest advances in clinical increasingly limited.” oncology research and also enriched for targets known to be associated with (or drive) childhood cancers John Bartlett, PhD Director of Transformative Pathology Platform • Based on robust Ion AmpliSeq™ technology, this assay Ontario Institute for Cancer Research -

Y-Box Binding Protein 1 Is Up-Regulated in Proliferative Breast Cancer and Its Inhibition Deregulates the Cell Cycle
483-492.qxd 21/6/2010 12:04 ÌÌ ™ÂÏ›‰·483 INTERNATIONAL JOURNAL OF ONCOLOGY 37: 483-492, 2010 483 Y-box binding protein 1 is up-regulated in proliferative breast cancer and its inhibition deregulates the cell cycle YING-NAN YU1, GEORGE WAI-CHEONG YIP1, PUAY-HOON TAN1,2, AYE AYE THIKE2, KEN MATSUMOTO3, MASAFUMI TSUJIMOTO3 and BOON-HUAT BAY1 1Department of Anatomy, Yong Loo Lin School of Medicine, National University of Singapore; 2Department of Pathology, Singapore General Hospital, Singapore; 3Laboratory of Cellular Biochemistry, Institute of Physical and Chemical Research (RIKEN), Wako, Japan Received March 4, 2010; Accepted April 27, 2010 DOI: 10.3892/ijo_00000697 Abstract. The Y-box-binding protein 1 (YB-1), a member of Introduction the cold-shock domain RNA-and DNA-binding protein family, has pleiotropic functions such as regulation of the Breast cancer is the most frequent malignancy among women cell cycle. The aim of this study was to evaluate if YB-1 is a in Western countries. It is estimated that more than 1.1 million proliferative marker in breast cancer and elucidate potential new breast cancer cases are diagnosed worldwide per year (1). downstream targets involved in YB-1-mediated cell cycle Identification of clinically useful biomarkers is imperative regulation using RNA interference technology. YB-1 protein for optimal care and management of breast tumors (2). Several expression was evaluated in tissue microarrays of 131 breast proteins such as estrogen receptors, epidermal growth factor invasive ductal carcinomas by immunohistochemistry, while receptors, BRCA1 and progesterone receptors have been the YB-1 gene expression profile was evaluated in the T-47D, reported to be prognostic biomarkers in breast cancer (3,4).