U5a1 Mitochondrial DNA Haplotype Identified in Eneolithic Skeleton from Shatar Chuluu, Mongolia
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mongolia Country Report 2018
Toxic Site Identification Program in Mongolia Award: DCI-ENV/2015/371157 Prepared by: Erdenesaikhan Naidansuren Prepared for: UNIDO Date: October 2018 Pure Earth 475 Riverside Drive, Suite 860 New York, NY, USA +1 212 647 8330 www.pureearth.org Acknowledgements ................................................................................................................. 3 Organizational Background .................................................................................................... 3 Toxic Site Identification Program (TSIP) ............................................................................... 3 Project Background ................................................................................................................. 5 Country Background ............................................................................................................... 5 Implimentation Strategy .......................................................................................................... 6 Coordinating with the Government ........................................................................................ 6 Sharing TSIP Information ....................................................................................................... 7 Current Work .......................................................................................................................... 8 TSIP Training in Mongolia ....................................................................................................... 9 Sites -

CBD Fifth National Report
CONVENTION ON CONVENTION ON BIOLOGICAL DIVERSITY BIOLOGICAL DIVERSITY THE 5TH NATIONAL REPORT OF MONGOLIA biolohJA JJa folea YeehcO beiide& oa KnWWn}A. T HE CONVENTION ON BIOLOGI 5 T H N A T IO N AL R EPO RT C AL DIVERSITY OF M O N GOLIA MINISTRY OF ENVIRONMENT AND GREEN DEVELOPMENT STEPPE FORWARD PROGRAMME, Government building II, BIOLOGY DEPARTMENT, United Nation’s street 5/2, NATIONAL UNIVERSITY OF MONGOLIA TH Chingeltei District, Ulaanbaatar 15160, NUM, Building-2, Ulaanbaatar, Mongolia THE 5 NATIONAL REPORT OF Mongolia P.O.Box 537, Ulaanbaatar 210646A, Tel: 976-51-266197 Ulaanbaatar, Mongolia E-mail: [email protected] Tel: 976-99180148; 976-88305909; 976-88083058 MONGOLIA E-mail: [email protected]; [email protected]; [email protected] Designed by Mongolica Publishing 2014 Ulaanbaatar, Mongolia. 2014 CONVENTION ON BIOLOGICAL DIVERSITY CONVENTION ON BIOLOGICAL DIVERSITY FINANCED BY: MINISTRY OF ENVIRONMENT AND GREEN DEVELOPMENT CONVENTION ON BIOLOGICAL DIVERSITY-MONGOLIA GLOBAL ENVIRONMENT FACILITY UNITED NATIONS ENVIRONMENTAL PROGRAM CONVENTION ON BIOLOGICAL DIVERSITY THE 5TH NATIONAL REPORT OF MONGOLIA REPORT COMPILERS: COMPILED BY: S. GOMBOBAATAR STEPPE FORWARD PROGRAMME, NUM S. MYAGMARSUREN N. CONABOY М. Мunkhjargal TAXON COMPILERS: PLANT: B. OYUNTSETSEG, M. URGAMAL INVERTEBRATE: S. GANTIGMAA Fish, aMphibian, reptile: kh. Тerbish BIRD: S. GOMBOBAATAR MAMMAL: S. SHAR CONTRIBUTIONS FROM: EDITORS: NATIONAL UNIVERSITY OF MONGOLIA INSTITUTE OF BIOLOGY, MONGOLIAN ACADEMY OF SCIENCES D. BATBOLD MONGOLIAN ORNITHOLOGICAL SOCIETY -

Remote Sensing
remote sensing Article Extreme Climate Event and Its Impact on Landscape Resilience in Gobi Region of Mongolia Oyudari Vova 1,* , Martin Kappas 1 , Tsolmon Renchin 2 and Steven R. Fassnacht 1,3,4,5 1 Cartography, GIS and Remote Sensing Department, Institute of Geography, University of Göttingen, 37007 Göttingen, Germany; [email protected] (M.K.); [email protected] (S.R.F.) 2 Department of Physics, National University of Mongolia, Ulaanbaatar 14200, Mongolia; [email protected] 3 Department of Ecosystem Science and Sustainability—Watershed Science, Colorado State University, Fort Collins, CO 80523-1476, USA 4 Cooperative Institute for Research in the Atmosphere, CSU, Fort Collins, CO 80523-1375, USA 5 Natural Resources Ecology Lab, Colorado State University, Fort Collins, CO 80523-1499, USA * Correspondence: [email protected]; Tel.: +49-176-25398594 Received: 9 July 2020; Accepted: 2 September 2020; Published: 5 September 2020 Abstract: The dzud, a specific type of climate disaster in Mongolia, is responsible for serious environmental and economic damage. It is characterized by heavy snowfall and severe winter conditions, causing mass livestock deaths that occur through the following spring. These events substantially limit socioeconomic development in Mongolia. In this research, we conducted an analysis of several dzud events (2000, 2001, 2002, and 2010) to understand the spatial and temporal variability of vegetation conditions in the Gobi region of Mongolia. The present paper also establishes how these extreme climatic events affect vegetation cover and local grazing conditions using the seasonal aridity index (aAIZ), time-series Moderate Resolution Imaging Spectroradiometer (MODIS) Normalized Difference Vegetation Index (NDVI), and livestock data. -

287 the Runic Inscription Monuments and Stamps in Bayankhongor
2nd International Congress On New Horizons In Education And Social Sciences (ICES-2019) Proceedings June 18-19, 2019 Istanbul-TURKEY The Runic inscription monuments and stamps in Bayankhongor province, Mongolia Assist.Prof. Dr. Gerelmaa. NAMSRAI1 Assist.Prof. Dr. Azzaya. BADAM2 Sen.lec. PhD (c). Otgonsuren. TSEDEN3 ABSTRACT In last two decades, numbers of runic inscriptions have been found in the western part of Mongolia: mostly in Bayan-Ulgii, Khovd and Govi-Altai provinces, and Pages: 287-295 comparatively less in Uvs and Zavkhan provinces, and also Bayankhongor province in which the following five inscriptions date back to the Ancient Turkic Period: Doi: 10.21733/ibad.585676 1. Inscription of Bömbögör monument (1986) 2. Inscription of the Three Mandalas: Adag mandala (the End Mandala) and Dund Mandala (The Middle Mandala) (1987) 3. Inscription of Olon Nuur (2008) 287 4. Inscription of Dalt (2012) 1 National University of Mongolia, 5. Inscription of Khirgisiin Ovoo Monument (2016). MONGOLIA, [email protected] Of them, the initial two inscriptions had been discovered by 1990, and the latter 2 National University of Mongolia, three since 2008, which have been followed by some alternative versions of MONGOLIA, [email protected] deciphering due to some inadequate reviews. So they have given us a motivation to review the meanings of the words and expressions incorporated in them, as the 3 National University of Mongolia, following: MONGOLIA, [email protected] These two inscriptions – properly viewed to be the classical monuments of the runic inscription, have some special words and expressions as well as personal names those are not noted in the monuments found in other provinces. -

Severe Winter 27 May 2010
Emergency appeal n° MDRMN004 Mongolia: GLIDE n° CW-2010-000010-MNG Operations update n° 2 Severe winter 27 May 2010 Period covered by this Ops Update: 23 April to 20 May 2010 Appeal target (current): CHF 1,062,295 Appeal coverage: 71% <click here to go directly to the updated donor response list, or here to contact details> Appeal history: • This Emergency Appeal was launched on 29 March 2010, seeking CHF 1,062,295 for six months period in order to assist 13,600 beneficiaries in 13 provinces whose vulnerability and needs have dramatically increased due to the seriousness of the climatic and social situation and its worsening. • Disaster Relief Emergency Fund (DREF): CHF 100,000 (USD 93,924 or EUR 70,006) was allocated from the Federation’s DREF to support the national society in this operation. Unearmarked funds to replenish DREF are encouraged. Mr. Ganbaatar’s family was entitled to food and non-food assistance from the DREF cold waves operation in March 2010 because the family has lost all the animals that belonged to them as well as those they were herding for a wage. In addition, the family has more than 3 children under 16 and mother is breastfeeding. During IFRC visit in April, it was reported that their food reserve will be enough until mid May and after that, the family will have no other solution to feed their children. Left without a single animal in the yard and having no alternative livelihood option, the parents were more than devastated about their children’s future but hide their emotions and worries to keep the family atmosphere warm and happy. -

Central Mongolia
© Lonely Planet Publications 101 Central Mongolia Roll out of Ulaanbaatar in a Russian jeep and you’ll only need to put a hill or two between yourself and the city before the vast steppes of cental Mongolia begin to unfold before your eyes. Verdant swaths of empty landscapes are sprinkled with tiny gers stretching to CENTRAL MONGOLIA the horizon while magical light plays through clouds and across the valleys. But central Mongolia offers more than steppes. Landscapes are broken by the forested hillsides of the Khan Khentii range, meandering rivers such as the Tuul and lunar-like lava fields spilling across central Arkhangai. The silhouette of a lone horseman on a hill or camels caravanning in the distance completes every perfect day. The rivers and back trails of Gorkhi-Terelj National Park beckon the outdoor enthusiast. At Khustain National Park you can break out the binoculars and spot the reintroduced takhi horse. Alternatively, set out from Ulaanbaatar on foot, climb the holy Bogdkhan Uul to the south of the city, and camp out by Mandshir Khiid. Travelling by horse is another great way to get around the region. Travellers with more time on their hands can spend weeks exploring the ancient sites and remote areas of the mighty Khangai and its surrounding plains. Central Mongolia’s aimags (provinces), Töv, Arkhangai and Övörkhangai, are the most vis- ited areas in the countryside. The roads and transport are far better here than in the rest of Mongolia, and there is plenty to see, including ancient monasteries, gorgeous lakes and many national parks. -

20101214 Mongolia Assessment DZUD Epc__KB Clean
2009/2010 MONGOLIA DZUD: DISPLACED RURAL HERDER COMMUNITIES RESPONSE ASSESSMENT AND INTENTIONS SURVEY (RAIS) FUNDED BY THE INTERNATIONAL ORGANIZATION FOR MIGRATION (IOM) AUGUST 2010 1 Table of Contents Table of Contents...............................................................................................................2 Glossary of Terms Executive Summary Recommendations for responses The Mongolian Migration Context.....................................................................................3 Dzud Affected Community Response Assessment and Intentions Survey (RAIS).............11 Analysis of Data from the RAIS.......................................................................................12 1. Impact of Dzud and Migrations............................................................................12 2. Perceived alternatives...........................................................................................17 3. Humanitarian Response........................................................................................18 4. Movement of Populations and Intentions..............................................................21 5. Access to Information...........................................................................................22 6. Summary of Data Analysis...................................................................................23 Annex 1 Areas and number of family surveyed................................................................25 Annex 2 Access DB Front Page.......................................................................................26 -

Mongolia 03M00 Iles NOVOSIBIRSK O
# e 0500km Mongolia 03m00 iles NOVOSIBIRSK O. Beloye Mana Bratsk Res. (Bratskoye Vdkhr.) Amarbayasgalant Khiid Gorkhi-Terelj RUSSIA Mongolia’s best-preserved National Park monastery Biking, hiking and horse- riding opportunities Ulaanbaatar Khövsgöl Nuur Premier horse-riding and Cosmopolitan capital with fishing area nightlife and museums RUSSIA Ölgii Lake Baikal Visit the Eagle Festival (Baygal Nuur) ^# Dadal #] Irkutsk Chita #\ Kyzyl On the trail of #] Ulan Ude Chinggis Khaan 33 #\ Renchinlkhumbe Khövsgöl Nuur Zaudinsky Uvs #\ National Park Border #÷ Crossing Ulaangom Nuur +# ^# Khatgal Tsagaan- Mongol Daguur Tsagaannuur 44UVS Züüngov #\ #\ ÷# #\ Border Siilkhem #– #\ –# Uur +# Strictly Protected 3333 333#\ Altanbulag 3 Bayanzürkh #\ Nuruu National Khan Khokhii Crossing Areas #\ Mǎnzhōulǐ Naranbulag ^# #\ #] Park #\ National Park KHÖVSGÖL #÷ ^# Ölgii Sükhbaatar Chuluunkhoroot ÷# Amarbayasgalant Hǎilā’ěr ÷# –# Tsagaan ^# Mörön BULGAN Onon-Balj Bayandun 44÷# #\ Uul #– Khiid National #\ Altai Tavan Khutag- Ú# #\ Darkhan BAYAN- Khyargas Nuur Tüdevtei #\ Park #\ Bogd33National 333333 #\ Shine- Öndör 333333333Bayan Uul3 ÖLGII National Park #÷ #\ Park Ider #\ ^# Bayangol Dadal Khovd #\ Erdenet #\ Batshireet Zavkhanmandal ^# #\ ^# 444 #\ #\ Khalkhiin ÷# Tosontsengel Ideriyn #\ Binder #– SELENGE Gorkhi-Terelj Choibalsan Gol Khar Us Nuur Tar ia t Bulgan ^# #\ ZAVKHAN #\ National Park #– National Park #÷ Village Khairkhan Batsumber Bayan- #\ 3333333#\ Uliastai 3333#\ 3 333#÷ 33333333333333Nömrög3 Strictly #\ Chandimani ULAANBAATAR Ovoo Bulgan -
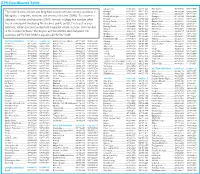
GPS Coordinates Table
GPS Coordinates Table Jiglegiin Am..................... 51º00.406 100º16.003 Mandalgov...................... 45º46.042 106º16.380 The table shows latitude and longitude coordinates for various locations in Khangal........................... 49º18.810 104º22.629 Mandal-Ovoo.................. 44º39.100 104º02.880 Khankh............................ 51º30.070 100º41.382 Manlai..............................44º04.441 106º51.703 Mongolia, in degrees, minutes and decimal minutes (DMM). To convert to Khar Bukh Balgas............. 47º53.198 103º53.513 Nomgon.......................... 42º50.160 105º08.983 degrees, minutes and seconds (DMS) format, multiply the number after Khatgal............................ 50º26.517 100º09.599 Ondorshil.........................45º13.585 108º15.223 Khishig-Öndör................. 48º17.678 103º27.086 Ongiin Khiid.....................45º20.367 104º00.306 the decimal point (including the decimal point) by 60. The result is your Khötöl..............................49º05.486 105º34.903 Orog Nuur....................... 45º02.692 100º36.314 seconds, which can be rounded to the nearest whole number. The minutes Khutag-Öndör................. 49º22.990 102º41.417 Saikhan-Ovoo..................45º27.459 103º54.110 Mogod.............................48º16.372 102º59.520 Sainshand.........................44º53.576 110º08.351 is the number between the degree symbol and the decimal point. For Mörön............................. 49º38.143 100º09.321 Sevrei...............................43º35.617 102º09.737 Orkhon........................... -

45145-001: Establishment of Climate-Resilient Rural Livelihoods
Initial Environmental Examination Project Number: 45145 December 2011 MON: Proposed Grant Assistance for Establishment of Climate-Resilient Rural Livelihoods (Financed by the Japan Fund for Poverty Reduction) Prepared by the Government of Mongolia for the Asian Development Bank. The initial environmental examination is a document of the borrower. The views expressed herein do not necessarily represent those of ADB’s Board of Directors, Management, or staff, and may be preliminary in nature. Your attention is directed to the “Terms of Use” section of this website. Initial Environmental Examination Establishment of Climate-Resilient Rural Livelihoods Prepared by the Government of Mongolia December 2011 CURRENCY EQUIVALENTS (as of 13 December 2011) Currency Unit – togrog (MNT) MNT1.00 = $0.00073 $1.00 = MNT1,379.00 ABBREVIATIONS ADB – Asian Development Bank EIA – environmental impact assessment EMP – environmental management plan HGO – herder group organization IEE – initial environmental examination JFPR – Japan Fund for Poverty Reduction MNET – Ministry of Nature, Environment and Tourism MOFALI – Ministry of Food, Agriculture and Light Industry PIU – project implementation unit PMU – project management unit SME – small and medium enterprise UNFCCC – United Nations Framework Convention for Climate Change WEIGHTS AND MEASURES ha – hectare km2 – square kilometer l – liter GLOSSARY aimag – province bagh – subdistrict soum – district NOTES (i) The fiscal year (FY) of the Government of Mongolia and its agencies ends on 31 December. FY before a calendar year denotes the year in which the fiscal year ends, e.g., FY2011 ends on 31 December 2011. (ii) In this report, “$” refers to US dollars. In preparing any country program or strategy, financing any project, or by making any designation of or reference to a particular territory or geographic area in this document, the Asian Development Bank does not intend to make any judgments as to the legal or other status of any territory or area. -

Mongolia: GLIDE N° CW-2010-000010-MNG Operations Update N° 2 Severe Winter 27 May 2010
Emergency appeal n° MDRMN004 Mongolia: GLIDE n° CW-2010-000010-MNG Operations update n° 2 Severe winter 27 May 2010 Period covered by this Ops Update: 23 April to 20 May 2010 Appeal target (current): CHF 1,062,295 Appeal coverage: 71% <click here to go directly to the updated donor response list, or here to contact details> Appeal history: This Emergency Appeal was launched on 29 March 2010, seeking CHF 1,062,295 for six months period in order to assist 13,600 beneficiaries in 13 provinces whose vulnerability and needs have dramatically increased due to the seriousness of the climatic and social situation and its worsening. Disaster Relief Emergency Fund (DREF): CHF 100,000 (USD 93,924 or EUR 70,006) was allocated from the Federation’s DREF to support the national society in this operation. Unearmarked funds to replenish DREF are encouraged. Mr. Ganbaatar’s family was entitled to food and non-food assistance from the DREF cold waves operation in March 2010 because the family has lost all the animals that belonged to them as well as those they were herding for a wage. In addition, the family has more than 3 children under 16 and mother is breastfeeding. During IFRC visit in April, it was reported that their food reserve will be enough until mid May and after that, the family will have no other solution to feed their children. Left without a single animal in the yard and having no alternative livelihood option, the parents were more than devastated about their children’s future but hide their emotions and worries to keep the family atmosphere warm and happy. -

論文 / 著書情報 Article / Book Information
論文 / 著書情報 Article / Book Information 題目(和文) Title(English) Study on the Influence of Interactive Learning Materials on Self- Regulated Learning for the Professional Development of Primary School Teachers in Mongolia 著者(和文) Li Shengru Author(English) Shengru Li 出典(和文) 学位:博士(工学), 学位授与機関:東京工業大学, 報告番号:甲第10966号, 授与年月日:2018年9月20日, 学位の種別:課程博士, 審査員:山口 しのぶ,髙田 潤一,阿部 直也,山下 幸彦,花岡 伸也 Citation(English) Degree:Doctor (Engineering), Conferring organization: Tokyo Institute of Technology, Report number:甲第10966号, Conferred date:2018/9/20, Degree Type:Course doctor, Examiner:,,,, 学位種別(和文) 博士論文 Type(English) Doctoral Thesis Powered by T2R2 (Tokyo Institute Research Repository) Study on the Influence of Interactive Learning Materials on Self-Regulated Learning for the Professional Development of Primary School Teachers in Mongolia By Shengru Li Yamaguchi-Takada Laboratory Submit in the partial fulfillment of the requirements for the degree of Doctor of Engineering in the Department of International Development Engineering Graduate School of Science and Engineering Tokyo Institute of Technology 2018 Table of Contents List of Tables ...................................................................................................... 6 Table of Figures .................................................................................................. 9 Abstract ............................................................................................................. 11 Thesis Summary ...............................................................................................