First Large-Scale DNA Barcoding Assessment of Reptiles in the Biodiversity Hotspot of Madagascar, Based on Newly Designed COI Primers
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

MADAGASCAR: the Wonders of the “8Th Continent” a Tropical Birding Custom Trip
MADAGASCAR: The Wonders of the “8th Continent” A Tropical Birding Custom Trip October 20—November 6, 2016 Guide: Ken Behrens All photos taken during this trip by Ken Behrens Annotated bird list by Jerry Connolly TOUR SUMMARY Madagascar has long been a core destination for Tropical Birding, and with the opening of a satellite office in the country several years ago, we further solidified our expertise in the “Eighth Continent.” This custom trip followed an itinerary similar to that of our main set-departure tour. Although this trip had a definite bird bias, it was really a general natural history tour. We took our time in observing and photographing whatever we could find, from lemurs to chameleons to bizarre invertebrates. Madagascar is rich in wonderful birds, and we enjoyed these to the fullest. But its mammals, reptiles, amphibians, and insects are just as wondrous and accessible, and a trip that ignored them would be sorely missing out. We also took time to enjoy the cultural riches of Madagascar, the small villages full of smiling children, the zebu carts which seem straight out of the Middle Ages, and the ingeniously engineered rice paddies. If you want to come to Madagascar and see it all… come with Tropical Birding! Madagascar is well known to pose some logistical challenges, especially in the form of the national airline Air Madagascar, but we enjoyed perfectly smooth sailing on this tour. We stayed in the most comfortable hotels available at each stop on the itinerary, including some that have just recently opened, and savored some remarkably good food, which many people rank as the best Madagascar Custom Tour October 20-November 6, 2016 they have ever had on any birding tour. -

The Herpetological Journal
Volume 3, Number 4 October 1993 ISSN 0268-0130 THE HERPETOLOGICAL JOURNAL Published by Indexed in THE BRITISH HERPETOLOGICAL SOCIETY Current Contents HERPETOLOGICAL JOURNAL, Vol. 3, pp. 124-129 SALMONELLOSIS DUE TO SALMONELLA HOUTEN IN CAPTIVE DAY GECKOS (GENUS: PHELSUMA GRAY) TRICHA J. MURPHY AND ALAN A. MYERS Departmenl of Zoology, Un iversity College, Cork, Ireland (A ccepted 19.11.92) ABSTRACT Two species of Sabnonella are reported fromcaptive Phelsuma spp., one of which (Sabnonella houten), was pathogenic. Twelve geckos developed clinical signs ofanorexia, diarrhoea, dehydration and cachexia. Ten died over a period of eight weeks and deaths occurred three to six weeks from the commencement of illness. Necropsy findings included dehydration, emaciation and liver necrosis. Sabnonella houten was isolated. The response of sick geckos to antibiotics and supportive therapy is discussed. INTRODUCTION have considered the routes of transmission of infection (Hinshaw & MacNeil, 1947; Collard & Montefiore, 1957; The normal flora of reptiles includes a wide range of Gram Chambon, Le Minor & Martin, 1959; Kaura & Singh, 1968; +VE and Gram -VE microorganisms especially of the family Kaura et al., 1970). Refai & Rohde (1969) and Sadek (1970) Enterobacteriaceae (Escherichia coli, Proteus spp., suggested transmission of Salmonella from mosquitoes and Klebsiella spp.), Aeromonas spp. and Pseudomonas spp. Bac other flies to geckos and from humans to reptiles (Iveson, teria also play an important role in reptilian diseases (Cooper, 1979). Pears of roonoses are not unfounded. A retrospective 1981). Potentially pathogenic organisms include Sabnonella survey of laboratory-confirmed cases of human clinical spp. of which over 1300 serotypes have been isolated from salmonellosis in the U. -

Blumgart Et Al 2017- Herpetological Survey Nosy Komba
Journal of Natural History ISSN: 0022-2933 (Print) 1464-5262 (Online) Journal homepage: http://www.tandfonline.com/loi/tnah20 Herpetological diversity across intact and modified habitats of Nosy Komba Island, Madagascar Dan Blumgart, Julia Dolhem & Christopher J. Raxworthy To cite this article: Dan Blumgart, Julia Dolhem & Christopher J. Raxworthy (2017): Herpetological diversity across intact and modified habitats of Nosy Komba Island, Madagascar, Journal of Natural History, DOI: 10.1080/00222933.2017.1287312 To link to this article: http://dx.doi.org/10.1080/00222933.2017.1287312 Published online: 28 Feb 2017. Submit your article to this journal Article views: 23 View related articles View Crossmark data Full Terms & Conditions of access and use can be found at http://www.tandfonline.com/action/journalInformation?journalCode=tnah20 Download by: [BBSRC] Date: 21 March 2017, At: 02:56 JOURNAL OF NATURAL HISTORY, 2017 http://dx.doi.org/10.1080/00222933.2017.1287312 Herpetological diversity across intact and modified habitats of Nosy Komba Island, Madagascar Dan Blumgart a, Julia Dolhema and Christopher J. Raxworthyb aMadagascar Research and Conservation Institute, BP 270, Hellville, Nosy Be, Madagascar; bDivision of Vertebrate Zoology, American, Museum of Natural History, New York, NY, USA ABSTRACT ARTICLE HISTORY A six month herpetological survey was undertaken between March Received 16 August 2016 and September 2015 on Nosy Komba, an island off of the north- Accepted 17 January 2017 west coast of mainland Madagascar which has undergone con- KEYWORDS fi siderable anthropogenic modi cation. A total of 14 species were Herpetofauna; conservation; found that have not been previously recorded on Nosy Komba, Madagascar; Nosy Komba; bringing the total island diversity to 52 (41 reptiles and 11 frogs). -

Download This PDF File
Check List 5(3): 692–694, 2009. ISSN: 1809-127X NOTES ON GEOGRAPHIC DISTRIBUTION Reptilia, Serpentes, Colubridae, Heteroliodon fohy: Distribution extension Steven Megson 1 Polly Mitchell 1 Neil D’Cruze 2 1 Frontier: The Society for Environmental Exploration. 50-52 Rivington Street. London, EC2A3QP, United Kingdom. 2 The World Society for the Protection of Animals. 89 Albert Embankment, London SE17TP, United Kingdom. E-mail: [email protected] Heteroliodon is an aglyphous, terrestrial and scale single, supralabials 7-7, third and fourth in poorly known snake genus endemic to contact with eye; dorsal scales smooth, in 17-17- Madagascar (Glaw and Vences 2007). 17 rows along body; preventrals 3, ventrals 133; Heteroliodon fohy can be identified by its dark anal plate divided; subcaudal pairs 58, a single brown dorsal coloration, whitish venter, yellowish spine at tail tip. This data shows that it closely nuchal band and whitish upper lip (Glaw et al. resembles the holotype. 2005). Its appearance is similar to Heteroliodon lava but the two can be readily distinguished as H. fohy has much fewer ventral scales (136 versus 214-224) (Nussbaum and Raxworthy 2000). Heteroliodon fohy was previously only known from the single type specimen found in the calcareous massif of Montagne des Français (Glaw et al. 2005; D’Cruze et al. 2007). On 25 October 2006 we found an adult specimen (total length 252 mm), in the region of Bobaomby in the extreme north of Madagascar, approximately 20 km north of the town of Antsiranana (Diego Suarez) and approximately 30 km north of the type locality in Montagne des Français (Figure 1). -

Ecosystem Profile Madagascar and Indian
ECOSYSTEM PROFILE MADAGASCAR AND INDIAN OCEAN ISLANDS FINAL VERSION DECEMBER 2014 This version of the Ecosystem Profile, based on the draft approved by the Donor Council of CEPF was finalized in December 2014 to include clearer maps and correct minor errors in Chapter 12 and Annexes Page i Prepared by: Conservation International - Madagascar Under the supervision of: Pierre Carret (CEPF) With technical support from: Moore Center for Science and Oceans - Conservation International Missouri Botanical Garden And support from the Regional Advisory Committee Léon Rajaobelina, Conservation International - Madagascar Richard Hughes, WWF – Western Indian Ocean Edmond Roger, Université d‘Antananarivo, Département de Biologie et Ecologie Végétales Christopher Holmes, WCS – Wildlife Conservation Society Steve Goodman, Vahatra Will Turner, Moore Center for Science and Oceans, Conservation International Ali Mohamed Soilihi, Point focal du FEM, Comores Xavier Luc Duval, Point focal du FEM, Maurice Maurice Loustau-Lalanne, Point focal du FEM, Seychelles Edmée Ralalaharisoa, Point focal du FEM, Madagascar Vikash Tatayah, Mauritian Wildlife Foundation Nirmal Jivan Shah, Nature Seychelles Andry Ralamboson Andriamanga, Alliance Voahary Gasy Idaroussi Hamadi, CNDD- Comores Luc Gigord - Conservatoire botanique du Mascarin, Réunion Claude-Anne Gauthier, Muséum National d‘Histoire Naturelle, Paris Jean-Paul Gaudechoux, Commission de l‘Océan Indien Drafted by the Ecosystem Profiling Team: Pierre Carret (CEPF) Harison Rabarison, Nirhy Rabibisoa, Setra Andriamanaitra, -

A Molecular Phylogeny of the Lamprophiidae Fitzinger (Serpentes, Caenophidia)
Zootaxa 1945: 51–66 (2008) ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ ZOOTAXA Copyright © 2008 · Magnolia Press ISSN 1175-5334 (online edition) Dissecting the major African snake radiation: a molecular phylogeny of the Lamprophiidae Fitzinger (Serpentes, Caenophidia) NICOLAS VIDAL1,10, WILLIAM R. BRANCH2, OLIVIER S.G. PAUWELS3,4, S. BLAIR HEDGES5, DONALD G. BROADLEY6, MICHAEL WINK7, CORINNE CRUAUD8, ULRICH JOGER9 & ZOLTÁN TAMÁS NAGY3 1UMR 7138, Systématique, Evolution, Adaptation, Département Systématique et Evolution, C. P. 26, Muséum National d’Histoire Naturelle, 43 Rue Cuvier, Paris 75005, France. E-mail: [email protected] 2Bayworld, P.O. Box 13147, Humewood 6013, South Africa. E-mail: [email protected] 3 Royal Belgian Institute of Natural Sciences, Rue Vautier 29, B-1000 Brussels, Belgium. E-mail: [email protected], [email protected] 4Smithsonian Institution, Center for Conservation Education and Sustainability, B.P. 48, Gamba, Gabon. 5Department of Biology, 208 Mueller Laboratory, Pennsylvania State University, University Park, PA 16802-5301 USA. E-mail: [email protected] 6Biodiversity Foundation for Africa, P.O. Box FM 730, Bulawayo, Zimbabwe. E-mail: [email protected] 7 Institute of Pharmacy and Molecular Biotechnology, University of Heidelberg, INF 364, D-69120 Heidelberg, Germany. E-mail: [email protected] 8Centre national de séquençage, Genoscope, 2 rue Gaston-Crémieux, CP5706, 91057 Evry cedex, France. E-mail: www.genoscope.fr 9Staatliches Naturhistorisches Museum, Pockelsstr. 10, 38106 Braunschweig, Germany. E-mail: [email protected] 10Corresponding author Abstract The Elapoidea includes the Elapidae and a large (~60 genera, 280 sp.) and mostly African (including Madagascar) radia- tion termed Lamprophiidae by Vidal et al. -

Evolution of the Iguanine Lizards (Sauria, Iguanidae) As Determined by Osteological and Myological Characters David F
Brigham Young University Science Bulletin, Biological Series Volume 12 | Number 3 Article 1 1-1971 Evolution of the iguanine lizards (Sauria, Iguanidae) as determined by osteological and myological characters David F. Avery Department of Biology, Southern Connecticut State College, New Haven, Connecticut Wilmer W. Tanner Department of Zoology, Brigham Young University, Provo, Utah Follow this and additional works at: https://scholarsarchive.byu.edu/byuscib Part of the Anatomy Commons, Botany Commons, Physiology Commons, and the Zoology Commons Recommended Citation Avery, David F. and Tanner, Wilmer W. (1971) "Evolution of the iguanine lizards (Sauria, Iguanidae) as determined by osteological and myological characters," Brigham Young University Science Bulletin, Biological Series: Vol. 12 : No. 3 , Article 1. Available at: https://scholarsarchive.byu.edu/byuscib/vol12/iss3/1 This Article is brought to you for free and open access by the Western North American Naturalist Publications at BYU ScholarsArchive. It has been accepted for inclusion in Brigham Young University Science Bulletin, Biological Series by an authorized editor of BYU ScholarsArchive. For more information, please contact [email protected], [email protected]. S-^' Brigham Young University f?!AR12j97d Science Bulletin \ EVOLUTION OF THE IGUANINE LIZARDS (SAURIA, IGUANIDAE) AS DETERMINED BY OSTEOLOGICAL AND MYOLOGICAL CHARACTERS by David F. Avery and Wilmer W. Tanner BIOLOGICAL SERIES — VOLUME Xil, NUMBER 3 JANUARY 1971 Brigham Young University Science Bulletin -

Print This Article
VOLUME 3 ISSUE 1 DECEMBER 2008 MADAGASCAR CONSERVATION & DEVELOPMENT INVESTING FOR A SUSTAINABLE NATURAL ENVIRONMENT FOR FUTURE GENERATIONS OF HUMANS, ANIMALS AND PLANTS OF MADAGASCAR IN THIS ISSUE Taboos & Social Contracts Bats & Bushmeat in Madagascar Endemic Plants in the Mandena Mining Area Radio for Sustain- able Development MADAGASCAR CONSERVATION & DEVELOPMENT VOLUME 3 | ISSUE 1 — DECEMBER 2008 PAGE 2 TABLE OF CONTENTS EDITORIAL 2 Editorial by Wilmé, L. and Waeber, P. O. 5 Foreword by Camara, C. Image in Action 85 Impressum The attachment that we feel to Madagascar compels us to talk ARTICLES about it – its richness, its values, its people and about life lessons 7 Taboos and social contracts: Tools for ecosystem learned and taught. As these experiences may differ in many management – lessons from the Manambolomaty aspects, a journal is the ideal place for sharing our common Lakes RAMSAR site, western Madagascar. Rabearivony J., ideas, as well as expressing our divergent thoughts and theories. Fanameha, E, Mampiandra, J. and Thorstom R. It is also a conduit for the exchange and transmission of our 17 Three flying fox (Pteropodidae: Pteropus rufus) ideas and perspectives to the world. Thus, it is the ambition roosts, three conservation challenges in southeastern of this journal to talk about Madagascar – it’s natural richness Madagascar. Rahaingodrahety, V. N., Andriafidison, D., and its conservation, about development and challenges in the Ratsimbazafy, J., Racey, P. A. and Jenkins, R. K. B. country, and more generally about components and facets of 22 Bats as bushmeat in Madagascar. Jenkins, R. K. B and conservation and development. Racey, P. -

Volume 2. Animals
AC20 Doc. 8.5 Annex (English only/Seulement en anglais/Únicamente en inglés) REVIEW OF SIGNIFICANT TRADE ANALYSIS OF TRADE TRENDS WITH NOTES ON THE CONSERVATION STATUS OF SELECTED SPECIES Volume 2. Animals Prepared for the CITES Animals Committee, CITES Secretariat by the United Nations Environment Programme World Conservation Monitoring Centre JANUARY 2004 AC20 Doc. 8.5 – p. 3 Prepared and produced by: UNEP World Conservation Monitoring Centre, Cambridge, UK UNEP WORLD CONSERVATION MONITORING CENTRE (UNEP-WCMC) www.unep-wcmc.org The UNEP World Conservation Monitoring Centre is the biodiversity assessment and policy implementation arm of the United Nations Environment Programme, the world’s foremost intergovernmental environmental organisation. UNEP-WCMC aims to help decision-makers recognise the value of biodiversity to people everywhere, and to apply this knowledge to all that they do. The Centre’s challenge is to transform complex data into policy-relevant information, to build tools and systems for analysis and integration, and to support the needs of nations and the international community as they engage in joint programmes of action. UNEP-WCMC provides objective, scientifically rigorous products and services that include ecosystem assessments, support for implementation of environmental agreements, regional and global biodiversity information, research on threats and impacts, and development of future scenarios for the living world. Prepared for: The CITES Secretariat, Geneva A contribution to UNEP - The United Nations Environment Programme Printed by: UNEP World Conservation Monitoring Centre 219 Huntingdon Road, Cambridge CB3 0DL, UK © Copyright: UNEP World Conservation Monitoring Centre/CITES Secretariat The contents of this report do not necessarily reflect the views or policies of UNEP or contributory organisations. -

Literature Cited in Lizards Natural History Database
Literature Cited in Lizards Natural History database Abdala, C. S., A. S. Quinteros, and R. E. Espinoza. 2008. Two new species of Liolaemus (Iguania: Liolaemidae) from the puna of northwestern Argentina. Herpetologica 64:458-471. Abdala, C. S., D. Baldo, R. A. Juárez, and R. E. Espinoza. 2016. The first parthenogenetic pleurodont Iguanian: a new all-female Liolaemus (Squamata: Liolaemidae) from western Argentina. Copeia 104:487-497. Abdala, C. S., J. C. Acosta, M. R. Cabrera, H. J. Villaviciencio, and J. Marinero. 2009. A new Andean Liolaemus of the L. montanus series (Squamata: Iguania: Liolaemidae) from western Argentina. South American Journal of Herpetology 4:91-102. Abdala, C. S., J. L. Acosta, J. C. Acosta, B. B. Alvarez, F. Arias, L. J. Avila, . S. M. Zalba. 2012. Categorización del estado de conservación de las lagartijas y anfisbenas de la República Argentina. Cuadernos de Herpetologia 26 (Suppl. 1):215-248. Abell, A. J. 1999. Male-female spacing patterns in the lizard, Sceloporus virgatus. Amphibia-Reptilia 20:185-194. Abts, M. L. 1987. Environment and variation in life history traits of the Chuckwalla, Sauromalus obesus. Ecological Monographs 57:215-232. Achaval, F., and A. Olmos. 2003. Anfibios y reptiles del Uruguay. Montevideo, Uruguay: Facultad de Ciencias. Achaval, F., and A. Olmos. 2007. Anfibio y reptiles del Uruguay, 3rd edn. Montevideo, Uruguay: Serie Fauna 1. Ackermann, T. 2006. Schreibers Glatkopfleguan Leiocephalus schreibersii. Munich, Germany: Natur und Tier. Ackley, J. W., P. J. Muelleman, R. E. Carter, R. W. Henderson, and R. Powell. 2009. A rapid assessment of herpetofaunal diversity in variously altered habitats on Dominica. -

FAUNE DE MADAGASCAR Publiée Sous Les Auspices Du Gouvernement De La République Malgache
FAUNE DE MADAGASCAR Publiée sous les auspices du Gouvernement de la République Malgache 47 REPTILES SAURIENS CHAMAELEONIDAE Genre Brookesia et complément pour le genre Chamae/eo par E.-R. BRYGûû (Mu.séUTn national dHistoire naturelle) Volume honoré d'une subvention de l'Agence de Coopération culturelle et technIque ÜR5TûM CNRS Paris 1978 FAUNE DE MADAGASCAR Collection fondée en 1956 par M. le Recteur Renaud PA LIAN Corre pondant de l'Institut Recteur de l'Académie de Bordeaux (alors Dirocteur adjoint de 1'1 RSM) Collection honorée d'une subvention de l'Académie des Scienoes (fonds Loutreuil) Comité de patronage M.le Dr RAIWTO RATSIMA~fANGA, membre correspondant de l'Institut, Paris. M.le Ministre de l1tducation nati nale, Tananarive. - M. le Président de l'Académie Malgache, Tananarive. - M. le Recteur de 1Université de Tananarive. - M. le Professeur de Zoologie de 1 niversité de Tananariv .- f. le DU'ecteur général du CNRS, Paris. - M. le Directeur général ct l üRSTüM, Pari. M. le Professeur Dr J. MILLOT, membre de l'ln titut, fondateur et ancien directeur de l'IRSM, Parjs. - M. Je Profe ur R. HEIM, fi mbre de lIn titut, Paris. MM. les Professeur J. DOR. T, membre de l'Institut, diJ'ecteul' du Muséum national, Paris; J.-M. PÉRÈS, membre de l'ln titut, Marseille; A. CILU3AUD, Paris; C. DELAMARE DEBouTTEVlLLE, Pari; P. LEHM ,Paris; M. RAKOTOMARIA, Tananarive. Comité de rédaction: M. R. PAlJLIA 1 Président; MM. C. DELAMARE DEBouTTEvILLE, P. DRACH, P. GRIVEA D, A. GRJEBINE, J.-J. PETTER, G. RAMANANTSOAVINA, P. ROEDERER, P. Vn:TTE ( ecrétaire). Les volumes de la «Faune de Madagascar », honorés d'une subvention de la République Malgache, sont publiés avec le concours financier du Centre National de la Recherche Scientifique et de l'Office de la Recherche Scientifique et Technique Outre-Mer. -
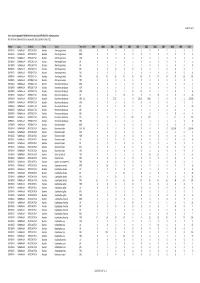
Gross Trade in Appendix II FAUNA (Direct Trade Only), 1999-2010 (For
AC25 Inf. 5 (1) Gross trade in Appendix II FAUNA (direct trade only), 1999‐2010 (for selection process) N.B. Data from 2009 and 2010 are incomplete. Data extracted 1 April 2011 Phylum Class TaxOrder Family Taxon Term Unit 1999 2000 2001 2002 2003 2004 2005 2006 2007 2008 2009 Total CHORDATA MAMMALIA ARTIODACTYLA Bovidae Ammotragus lervia BOD 0 00001000102 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Ammotragus lervia BON 0 00080000008 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Ammotragus lervia HOR 0 00000110406 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Ammotragus lervia LIV 0 00060000006 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Ammotragus lervia SKI 1 11311000008 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Ammotragus lervia SKP 0 00000010001 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Ammotragus lervia SKU 2 052101000011 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Ammotragus lervia TRO 15 42 49 43 46 46 27 27 14 37 26 372 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Antilope cervicapra TRO 0 00000020002 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Bison bison athabascae BOD 0 00100001002 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Bison bison athabascae HOP 0 00200000002 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Bison bison athabascae HOR 0 0010100120216 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Bison bison athabascae LIV 0 0 5 14 0 0 0 30 0 0 0 49 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Bison bison athabascae MEA KIL 0 5 27.22 0 0 272.16 1000 00001304.38 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Bison bison athabascae MEA 0 00000000101 CHORDATA MAMMALIA ARTIODACTYLA Bovidae Bison bison athabascae