Modified Nucleic Acids: Expanding the Capabilities of Functional
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

(12) Patent Application Publication (10) Pub. No.: US 2013/0203610 A1 Meller Et Al
US 20130203610A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2013/0203610 A1 Meller et al. (43) Pub. Date: Aug. 8, 2013 (54) TOOLS AND METHOD FOR NANOPORES Related U.S. Application Data UNZIPPING-DEPENDENT NUCLECACID (60) Provisional application No. 61/318,872, filed on Mar. SEQUENCING 30, 2010. (75) Inventors: Amit Meller, Brookline, MA (US); Alon Publication Classification Singer, Brighton, MA (US) (51) Int. Cl. (73) Assignee: TRUSTEES OF BOSTON CI2O I/68 (2006.01) UNIVERSITY, Boston, MA (US) (52) U.S. Cl. CPC .................................... CI2O I/6874 (2013.01) (21) Appl. No.: 13/638,455 USPC ................................................. 506/6:506/16 (57) ABSTRACT (22) PCT Filed: Mar. 30, 2011 Provided herein is a library that comprises a plurality of molecular beacons (MBs), each MB having a detectable (86). PCT No.: PCT/US2O11AO3O430 label, a detectable label blocker and a modifier group. The S371 (c)(1), library is used in conjunction with nanopore unzipping-de (2), (4) Date: Apr. 17, 2013 pendent sequencing of nucleic acids. Patent Application Publication Aug. 8, 2013 Sheet 1 of 18 US 2013/020361.0 A1 . N s Patent Application Publication Aug. 8, 2013 Sheet 2 of 18 US 2013/020361.0 A1 I’9IAI ::::::::::: Dº3.modoueN Patent Application Publication Aug. 8, 2013 Sheet 3 of 18 US 2013/020361.0 A1 Z’9IAI ~~~~~~~~~);........ Patent Application Publication Aug. 8, 2013 Sheet 4 of 18 US 2013/020361.0 A1 s :·.{-zzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzzz iii. 9 iii.338 lii &S Patent Application Publication Aug. 8, 2013 Sheet 5 of 18 US 2013/020361.0 A1 s r Patent Application Publication Aug. -

Sequence-Selective Recognition of Double-Stranded RNA And
Downloaded from rnajournal.cshlp.org on October 8, 2021 - Published by Cold Spring Harbor Laboratory Press Hnedzko et al. Sequence-Selective Recognition of Double-Stranded RNA and Enhanced Cellular Uptake of Cationic Nucleobase and Backbone- Modified Peptide Nucleic Acids Dziyana Hnedzko1,*, Dennis W. McGee,2 Yannis A. Karamitas1 and Eriks Rozners1,* Departments of 1 Chemistry and 2 Biological Sciences, Binghamton University, The State University of New York, Binghamton, New York 13902, United States. * To whom correspondence should be addressed. Tel: +1-607-777-2441; Fax: +1-607-777- 4478; Email: [email protected] and [email protected] Running Tittle: RNA recognition and cellular uptake of PNA 1 Downloaded from rnajournal.cshlp.org on October 8, 2021 - Published by Cold Spring Harbor Laboratory Press Hnedzko et al. ABSTRACT Sequence-selective recognition of complex RNAs in live cells could find broad applications in biology, biomedical research and biotechnology. However, specific recognition of structured RNA is challenging and generally applicable and effective methods are lacking. Recently, we found that peptide nucleic acids (PNAs) were unusually well suited ligands for recognition of double-stranded RNAs. Herein, we report that 2-aminopyridine (M) modified PNAs and their conjugates with lysine and arginine tripeptides form strong (Ka = 9.4 to 17 × 107 M-1) and sequence-selective triple helices with RNA hairpins at physiological pH and salt concentration. The affinity of PNA-peptide conjugates for the matched RNA hairpins was unusually high compared to the much lower affinity for DNA hairpins of the same 7 -1 sequence (Ka = 0.05 to 0.11 × 10 M ). The binding of double-stranded RNA by M-modified 4 -1 -1 PNA-peptide conjugates was a relatively fast process (kon = 2.9 × 10 M s ) compared to 3 -1 -1 the notoriously slow triple helix formation by oligodeoxynucleotides (kon ~ 10 M s ). -
Eukaryotic Non-Coding Rnas: New Targets for Diagnostics and Therapeutics?
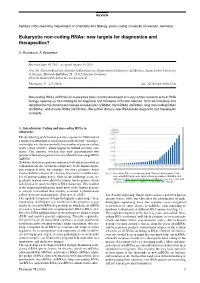
REVIEW Institute of Biochemistry, Department of Chemistry and Biology, Justus Liebig University of Giessen, Germany Eukaryotic non-coding RNAs: new targets for diagnostics and therapeutics? O. Rossbach, A. Bindereif Received June 30, 2015, accepted August 19, 2015 Prof. Dr. Albrecht Bindereif, Institute of Biochemistry, Department of Chemistry and Biology, Justus Liebig University of Giessen, Heinrich-Buff-Ring 58, 35392 Giessen, Germany [email protected] Pharmazie 71: 3–7 (2016) doi: 10.1691/ph.2016.5736 Non-coding RNAs (ncRNAs) in eukaryotes have recently developed to a very active research area in RNA biology, opening up new strategies for diagnosis and therapies of human disease. Here we introduce and describe the most important classes of eukaryotic ncRNAs: microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs). We further discuss new RNA-based diagnostic and therapeutic concepts. 1. Introduction: Coding and non-coding RNAs in eukaryotes The deciphering of the human genome sequence in 2000 marked a unique breakthrough in modern molecular biology. An impor- tant insight was the unexpectedly low number of protein-coding genes (about 20,000), which lagged far behind previous esti- mates. This number revealed that only approximately two percent of the human genome is transcribed into messenger RNA (mRNA). However, the human genome sequence itself did not provide an explanation for the enormous complexity of the human organ- ism compared with, for example, the more primitive worm Caenorhabditis elegans (C. elegans) that carries a similar num- Fig. 1: Non-coding RNAs as an emerging field. The rapid development of the ber of protein-coding genes. -

Deoxyguanosine Cytotoxicity by a Novel Inhibitor of Furine Nucleoside Phosphorylase, 8-Amino-9-Benzylguanine1
[CANCER RESEARCH 46, 519-523, February 1986] Potentiation of 2'-Deoxyguanosine Cytotoxicity by a Novel Inhibitor of Furine Nucleoside Phosphorylase, 8-Amino-9-benzylguanine1 Donna S. Shewach,2 Ji-Wang Chern, Katherine E. Pillóte,Leroy B. Townsend, and Peter E. Daddona3 Departments of Internal Medicine [D.S.S., P.E.D.], Biological Chemistry [P.E.D.], and Medicinal Chemistry [J-W.C., K.E.P., L.B.T.], University ol Michigan, Ann Arbor, Michigan 48109 ABSTRACT to the ADA-deficient disease state (2). PNP is an essential enzyme of the purine salvage pathway, We have synthesized and evaluated a series of 9-substituted catalyzing the phosphorolysis of guanosine, inosine, and their analogues of 8-aminoguanine, a known inhibitor of human purine 2'-deoxyribonucleoside derivatives to the respective purine nucleoside phosphorylase (PNP) activity. The ability of these bases. To date, several inhibitors of PNP have been identified, agents to inhibit PNP has been investigated. All compounds were and most of these compounds resemble purine bases or nucleo found to act as competitive (with inosine) inhibitors of PNP, with sides. The most potent inhibitors exhibit apparent K¡values in K¡values ranging from 0.2 to 290 /¿M.Themost potent of these the range of 10~6to 10~7 M (9-12). Using partially purified human analogues, 8-amino-9-benzylguanine, exhibited a K, value that erythrocyte PNP, the diphosphate derivative of acyclovir dis was 4-fold lower than that determined for the parent base, 8- played K¡values of 5.1 x 10~7 to 8.7 x 10~9 M, depending on aminoguanine. -

2'-Deoxyguanosine Toxicity for B and Mature T Lymphoid Cell Lines Is Mediated by Guanine Ribonucleotide Accumulation
2'-deoxyguanosine toxicity for B and mature T lymphoid cell lines is mediated by guanine ribonucleotide accumulation. Y Sidi, B S Mitchell J Clin Invest. 1984;74(5):1640-1648. https://doi.org/10.1172/JCI111580. Research Article Inherited deficiency of the enzyme purine nucleoside phosphorylase (PNP) results in selective and severe T lymphocyte depletion which is mediated by its substrate, 2'-deoxyguanosine. This observation provides a rationale for the use of PNP inhibitors as selective T cell immunosuppressive agents. We have studied the relative effects of the PNP inhibitor 8- aminoguanosine on the metabolism and growth of lymphoid cell lines of T and B cell origin. We have found that 2'- deoxyguanosine toxicity for T lymphoblasts is markedly potentiated by 8-aminoguanosine and is mediated by the accumulation of deoxyguanosine triphosphate. In contrast, the growth of T4+ mature T cell lines and B lymphoblast cell lines is inhibited by somewhat higher concentrations of 2'-deoxyguanosine (ID50 20 and 18 microM, respectively) in the presence of 8-aminoguanosine without an increase in deoxyguanosine triphosphate levels. Cytotoxicity correlates instead with a three- to fivefold increase in guanosine triphosphate (GTP) levels after 24 h. Accumulation of GTP and growth inhibition also result from exposure to guanosine, but not to guanine at equimolar concentrations. B lymphoblasts which are deficient in the purine salvage enzyme hypoxanthine guanine phosphoribosyltransferase are completely resistant to 2'-deoxyguanosine or guanosine concentrations up to 800 microM and do not demonstrate an increase in GTP levels. Growth inhibition and GTP accumulation are prevented by hypoxanthine or adenine, but not by 2'-deoxycytidine. -

Alternative Biochemistries for Alien Life: Basic Concepts and Requirements for the Design of a Robust Biocontainment System in Genetic Isolation
G C A T T A C G G C A T genes Review Alternative Biochemistries for Alien Life: Basic Concepts and Requirements for the Design of a Robust Biocontainment System in Genetic Isolation Christian Diwo 1 and Nediljko Budisa 1,2,* 1 Institut für Chemie, Technische Universität Berlin Müller-Breslau-Straße 10, 10623 Berlin, Germany; [email protected] 2 Department of Chemistry, University of Manitoba, 144 Dysart Rd, 360 Parker Building, Winnipeg, MB R3T 2N2, Canada * Correspondence: [email protected] or [email protected]; Tel.: +49-30-314-28821 or +1-204-474-9178 Received: 27 November 2018; Accepted: 21 December 2018; Published: 28 December 2018 Abstract: The universal genetic code, which is the foundation of cellular organization for almost all organisms, has fostered the exchange of genetic information from very different paths of evolution. The result of this communication network of potentially beneficial traits can be observed as modern biodiversity. Today, the genetic modification techniques of synthetic biology allow for the design of specialized organisms and their employment as tools, creating an artificial biodiversity based on the same universal genetic code. As there is no natural barrier towards the proliferation of genetic information which confers an advantage for a certain species, the naturally evolved genetic pool could be irreversibly altered if modified genetic information is exchanged. We argue that an alien genetic code which is incompatible with nature is likely to assure the inhibition of all mechanisms of genetic information transfer in an open environment. The two conceivable routes to synthetic life are either de novo cellular design or the successive alienation of a complex biological organism through laboratory evolution. -

Nuclear and Mitochondrial DNA Sequences from Two Denisovan Individuals
Nuclear and mitochondrial DNA sequences from two Denisovan individuals Susanna Sawyera,1, Gabriel Renauda,1, Bence Violab,c,d, Jean-Jacques Hublinc, Marie-Theres Gansaugea, Michael V. Shunkovd,e, Anatoly P. Dereviankod,f, Kay Prüfera, Janet Kelsoa, and Svante Pääboa,2 aDepartment of Evolutionary Genetics, Max Planck Institute for Evolutionary Anthropology, D-04103 Leipzig, Germany; bDepartment of Anthropology, University of Toronto, Toronto, ON M5S 2S2, Canada; cDepartment of Human Evolution, Max Planck Institute for Evolutionary Anthropology, D-04103 Leipzig, Germany; dInstitute of Archaeology and Ethnography, Russian Academy of Sciences, Novosibirsk, RU-630090, Russia; eNovosibirsk National Research State University, Novosibirsk, RU-630090, Russia; and fAltai State University, Barnaul, RU-656049, Russia Contributed by Svante Pääbo, October 13, 2015 (sent for review April 16, 2015; reviewed by Hendrik N. Poinar, Fred H. Smith, and Chris B. Stringer) Denisovans, a sister group of Neandertals, have been described on DNA to the ancestors of present-day populations across Asia the basis of a nuclear genome sequence from a finger phalanx and Oceania suggests that in addition to the Altai Mountains, (Denisova 3) found in Denisova Cave in the Altai Mountains. The they may have lived in other parts of Asia. In addition to the only other Denisovan specimen described to date is a molar (Deni- finger phalanx, a molar (Denisova 4) was found in the cave in sova 4) found at the same site. This tooth carries a mtDNA se- 2000. Although less than 0.2% of the DNA in the tooth derives quence similar to that of Denisova 3. Here we present nuclear from a hominin source, the mtDNA was sequenced and differed DNA sequences from Denisova 4 and a morphological description, from the finger phalanx mtDNA at only two positions, suggesting as well as mitochondrial and nuclear DNA sequence data, from it too may be from a Denisovan (2, 3). -

Evaluation of Locked Nucleic Acid–Modified Small Interfering RNA in Vitro and in Vivo
833 Evaluation of locked nucleic acid–modified small interfering RNA in vitro and in vivo Olaf R. Mook, Frank Baas, Marit B. de Wissel, model. Therefore, LNA-modified siRNA should be pre- and Kees Fluiter ferred over unmodified siRNA. [Mol Cancer Ther 2007; 6(3):833–43] Department of Neurogenetics, Academic Medical Center, Amsterdam, the Netherlands Introduction RNA interference (RNAi) is a natural process that affects Abstract gene silencing in eukaryotic systems at transcriptional, RNA interference has become widely used as an experi- posttranscriptional, and/or translational levels (1). Small mental tool to study gene function. In addition, small interfering RNA (siRNA) molecules are the key intermedi- interfering RNA (siRNA) may have great potential for the ates in this process, which can potentially inhibit the treatment of diseases. Recently, it was shown that siRNA expression of any given target gene. siRNA molecules hold can be used to mediate gene silencing in mouse models. great promise as biological tools and as potential thera- Locally administered siRNAs entered the first clinical trials, peutic agents for targeted inhibition of disease-causing but strategies for successful systemic delivery of siRNA genes. are still under development. Challenges still exist about the To optimize the use of siRNA as a therapeutic therapy, stability, delivery, and therapeutic efficacy of siRNA. In several modes of delivery have been tested in vivo. the present study, we compare the efficacy of two Improved delivery in animals has been achieved by methods of systemic siRNA delivery and the effects of complexation with cationic liposomes (2), polyethyleni- siRNA modifications using locked nucleic acids (LNA) in a mine (3), and Arg-Gly-Asp–polyethylene glycol–polyethy- xenograft cancer model. -

A Pirna-Like Small RNA Interacts with and Modulates P-ERM Proteins in Human Somatic Cells
ARTICLE Received 18 Mar 2015 | Accepted 27 Apr 2015 | Published 22 June 2015 DOI: 10.1038/ncomms8316 OPEN A piRNA-like small RNA interacts with and modulates p-ERM proteins in human somatic cells Yuping Mei1, Yuyan Wang1,2, Priti Kumari3, Amol Carl Shetty3, David Clark1, Tyler Gable1, Alexander D. MacKerell4,5, Mark Z. Ma1, David J. Weber5,6, Austin J. Yang5, Martin J. Edelman5 & Li Mao1,5 PIWI-interacting RNAs (piRNAs) are thought to silence transposon and gene expression during development. However, the roles of piRNAs in somatic tissues are largely unknown. Here we report the identification of 555 piRNAs in human lung bronchial epithelial (HBE) and non-small cell lung cancer (NSCLC) cell lines, including 295 that do not exist in databases termed as piRNA-like sncRNAs or piRNA-Ls. Distinctive piRNA/piRNA-L expression patterns are observed between HBE and NSCLC cells. piRNA-like-163 (piR-L-163), the top down- regulated piRNA-L in NSCLC cells, binds directly to phosphorylated ERM proteins (p-ERM), which is dependent on the central part of UUNNUUUNNUU motif in piR-L-163 and the RRRKPDT element in ERM. The piR-L-163/p-ERM interaction is critical for p-ERM’s binding capability to filamentous actin (F-actin) and ERM-binding phosphoprotein 50 (EBP50). Thus, piRNA/piRNA-L may play a regulatory role through direct interaction with proteins in physiological and pathophysiological conditions. 1 Department of Oncology and Diagnostic Sciences, University of Maryland School of Dentistry, 650W Baltimore Street, Baltimore, Maryland 21201, USA. 2 Department of Thoracic Medical Oncology, Peking University Cancer Hospital and Institute, Beijing 100142, China. -

Locked Nucleic Acid
lysi ana s & io B B io f m o e l d a Journal of Mishra and Mukhopadhyay, J Bioanal Biomed 2013, 5:2 i n c r i n u e DOI: 10.4172/1948-593X.1000e114 o J ISSN: 1948-593X Bioanalysis & Biomedicine Editorial OpenOpen Access Access Locked Nucleic Acid (LNA)-based Nucleic Acid Sensors Sourav Mishra and Rupa Mukhopadhyay* Department of Biological Chemistry, Indian Association for the Cultivation of Science, Jadavpur, Kolkata-700 032, India In recent times, much developments in the field of ‘nucleic acid may prevent interactions with the solid substrates, [15] and it can have sensors’, especially using alternative nucleic acid probes like peptide multiple water bridges that provide it with extra stability compared to nucleic acid (PNA) and locked nucleic acid (LNA), have taken place. DNA or RNA. It has been shown that both the highest Tm increase Although most of these are concerned with ‘solution phase’ studies, per LNA modification and the best mismatch discrimination are some reports have been made on ‘on-surface’ detection. The latter type achieved for short LNA sequences [16]. In addition to that, LNA of detection is particularly important to nurture, considering clinical phosphoramidites and their oligomers are commercially available, diagnostic applications using microarrays. In this article, we’ll briefly and LNA nucleotides can be mixed with those of the natural nucleic present the primary developments reported in the past two decades, acids for generating heterogeneous probe molecules. These properties along with possibilities for future developments, in case of the LNA- of LNA hold the promise that it can be a potentially better alternative based sensors. -

Hammerhead Ribozymes Against Virus and Viroid Rnas
Hammerhead Ribozymes Against Virus and Viroid RNAs Alberto Carbonell, Ricardo Flores, and Selma Gago Contents 1 A Historical Overview: Hammerhead Ribozymes in Their Natural Context ................................................................... 412 2 Manipulating Cis-Acting Hammerheads to Act in Trans ................................. 414 3 A Critical Issue: Colocalization of Ribozyme and Substrate . .. .. ... .. .. .. .. .. ... .. .. .. .. 416 4 An Unanticipated Participant: Interactions Between Peripheral Loops of Natural Hammerheads Greatly Increase Their Self-Cleavage Activity ........................... 417 5 A New Generation of Trans-Acting Hammerheads Operating In Vitro and In Vivo at Physiological Concentrations of Magnesium . ...... 419 6 Trans-Cleavage In Vitro of Short RNA Substrates by Discontinuous and Extended Hammerheads ........................................... 420 7 Trans-Cleavage In Vitro of a Highly Structured RNA by Discontinuous and Extended Hammerheads ........................................... 421 8 Trans-Cleavage In Vivo of a Viroid RNA by an Extended PLMVd-Derived Hammerhead ........................................... 422 9 Concluding Remarks and Outlooks ........................................................ 424 References ....................................................................................... 425 Abstract The hammerhead ribozyme, a small catalytic motif that promotes self- cleavage of the RNAs in which it is found naturally embedded, can be manipulated to recognize and cleave specifically -

Central Nervous System Dysfunction and Erythrocyte Guanosine Triphosphate Depletion in Purine Nucleoside Phosphorylase Deficiency
Arch Dis Child: first published as 10.1136/adc.62.4.385 on 1 April 1987. Downloaded from Archives of Disease in Childhood, 1987, 62, 385-391 Central nervous system dysfunction and erythrocyte guanosine triphosphate depletion in purine nucleoside phosphorylase deficiency H A SIMMONDS, L D FAIRBANKS, G S MORRIS, G MORGAN, A R WATSON, P TIMMS, AND B SINGH Purine Laboratory, Guy's Hospital, London, Department of Immunology, Institute of Child Health, London, Department of Paediatrics, City Hospital, Nottingham, Department of Paediatrics and Chemical Pathology, National Guard King Khalid Hospital, Jeddah, Saudi Arabia SUMMARY Developmental retardation was a prominent clinical feature in six infants from three kindreds deficient in the enzyme purine nucleoside phosphorylase (PNP) and was present before development of T cell immunodeficiency. Guanosine triphosphate (GTP) depletion was noted in the erythrocytes of all surviving homozygotes and was of equivalent magnitude to that found in the Lesch-Nyhan syndrome (complete hypoxanthine-guanine phosphoribosyltransferase (HGPRT) deficiency). The similarity between the neurological complications in both disorders that the two major clinical consequences of complete PNP deficiency have differing indicates copyright. aetiologies: (1) neurological effects resulting from deficiency of the PNP enzyme products, which are the substrates for HGPRT, leading to functional deficiency of this enzyme. (2) immunodeficiency caused by accumulation of the PNP enzyme substrates, one of which, deoxyguanosine, is toxic to T cells. These studies show the need to consider PNP deficiency (suggested by the finding of hypouricaemia) in patients with neurological dysfunction, as well as in T cell immunodeficiency. http://adc.bmj.com/ They suggest an important role for GTP in normal central nervous system function.