Supplementary Figures and Tables
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Keratin 19 Regulates Cell Shape and Cell-Cell Adhesion of MCF7 Cells While Maintaining E-Cadherin Localization at the Cell Surface
Keratin 19 regulates cell shape and cell-cell adhesion of MCF7 cells while maintaining E-cadherin localization at the cell surface Welcome to my poster. This is Sarah Alsharif, a PhD student from the biology department. I am glad to present the work our lab has been doing in the breast cancer field. In fact, after lung cancer, breast cancer is the second cause of death in women worldwide (1). It is estimated that every 18 seconds, approximately one new case of breast cancer is documented (2). No one dies due to cancer itself. The death is because of metastasis which takes place when cancer cells leave a breast in which they are formed and reach other sites such as the brain or lung. Our lab is interested in investigating the mechanism behind metastasis of breast cancer. Metastasis is associated with what is called epithelial to mesenchymal transition (EMT), the process characterized by loss of cell to cell adhesion and expression of epithelial markers such as keratin intermediate filament proteins, as you can see in the first three images of cells. Those filaments are keratins and they are critical for the shape and for maintaining mechanical integrity of epithelial cells via cell to cell complexes called desmosomes. Among different keratins, keratin 19 (K19) is highly expressed in many types of cancer including breast cancer, and is correlated with a worse prognosis (3). Consistently, K19 expression has been reported to be significantly higher in metastatic breast cancer tumor cells compared to primary tumors (4). The role of K19 on mechanical properties of cancer cells for cell migration and possible impact on metastasis in breast cancer patients is still unknown. -

Kinesin-14 Motor Protein KIFC1 Participates in DNA Synthesis and Chromatin Maintenance Ya-Lan Wei1 and Wan-Xi Yang 1
Wei and Yang Cell Death and Disease (2019) 10:402 https://doi.org/10.1038/s41419-019-1619-9 Cell Death & Disease ARTICLE Open Access Kinesin-14 motor protein KIFC1 participates in DNA synthesis and chromatin maintenance Ya-Lan Wei1 and Wan-Xi Yang 1 Abstract The nuclear localization signal (NLS) in kinesin-14 KIFC1 is associated with nuclear importins and Ran gradient, but detailed mechanism remains unknown. In this study, we found that KIFC1 proteins have specific transport characteristics during cell cycle. In the absence of KIFC1, cell cycle kinetics decrease significantly with a prolonged S phase. After KIFC1 overexpression, the duration of S phase becomes shorten. KIFC1 may transport the recombinant/ replicate-related proteins into the nucleus, meanwhile avoiding excessive KIFC1 in the cytoplasm, which results in aberrant microtubule bundling. Interestingly, the deletion of kifc1 in human cells results in a higher ratio of aberrant nuclear membrane, and the degradation of lamin B and lamin A/C. We also found that kifc1 deletion leads to defects in metaphase mitotic spindle assembly, and then results in chromosome structural abnormality. The kifc1-/- cells finally form micronuclei in daughter cells, and results in aneuploidy and chromosome loss in cell cycle. In this study, we demonstrate that kinesin-14 KIFC1 proteins involve in regulating DNA synthesis in S phase, and chromatin maintenance in mitosis, and maintain cell growth in a nuclear transport-independent way. 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; Introduction KIFC1 mainly cluster the spindles involving in chromo- Kinesin-14 KIFC1 transports various cargos along the some alignment and segregation. While chromokinesins microtubule to the minus ends1. -

Inhibiting TNIK for Treating Colon Cancer
(19) & (11) EP 2 305 717 A1 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 06.04.2011 Bulletin 2011/14 C07K 16/40 (2006.01) C12N 15/11 (2006.01) C12Q 1/48 (2006.01) C12Q 1/68 (2006.01) (2006.01) (21) Application number: 09170853.7 G01N 33/50 (22) Date of filing: 21.09.2009 (84) Designated Contracting States: • Mahmoudi, Tokameh AT BE BG CH CY CZ DE DK EE ES FI FR GB GR 3515 XS, Utrecht (NL) HR HU IE IS IT LI LT LU LV MC MK MT NL NO PL • Clevers, Johannes Carolus PT RO SE SI SK SM TR 3712 AP, Huis ter Heide (NL) (71) Applicant: KoninklijkeNederlandse Akademie van (74) Representative: Swinkels, Bart Willem Wetenschappen Nederlandsch Octrooibureau 1011 JV Amsterdam (NL) J. W. Frisolaan 13 2517 JS Den Haag (NL) (72) Inventors: • Wing Li, Vivian Sze 3572 SH, Utrecht (NL) (54) Inhibiting TNIK for treating colon cancer (57) The invention relates to an inhibitor of TNIK and its use for treating cancer. EP 2 305 717 A1 Printed by Jouve, 75001 PARIS (FR) EP 2 305 717 A1 Description Field of the invention 5 [0001] The invention relates to an inhibitor of TNIK and its use as a medicament for treating cancer. Background of the invention [0002] The primary function of the intestinal tract involves the digestion and absorption of nutrients. The intestinal 10 lumen is lined with a specialized simple epithelium, which performs the primary functions of digestion, water and nutrient absorption and forms a barrier against luminal pathogens. -

Viewed Under 23 (B) Or 203 (C) fi M M Male Cko Mice, and Largely Unaffected Magni Cation; Scale Bars, 500 M (B) and 50 M (C)
BRIEF COMMUNICATION www.jasn.org Renal Fanconi Syndrome and Hypophosphatemic Rickets in the Absence of Xenotropic and Polytropic Retroviral Receptor in the Nephron Camille Ansermet,* Matthias B. Moor,* Gabriel Centeno,* Muriel Auberson,* † † ‡ Dorothy Zhang Hu, Roland Baron, Svetlana Nikolaeva,* Barbara Haenzi,* | Natalya Katanaeva,* Ivan Gautschi,* Vladimir Katanaev,*§ Samuel Rotman, Robert Koesters,¶ †† Laurent Schild,* Sylvain Pradervand,** Olivier Bonny,* and Dmitri Firsov* BRIEF COMMUNICATION *Department of Pharmacology and Toxicology and **Genomic Technologies Facility, University of Lausanne, Lausanne, Switzerland; †Department of Oral Medicine, Infection, and Immunity, Harvard School of Dental Medicine, Boston, Massachusetts; ‡Institute of Evolutionary Physiology and Biochemistry, St. Petersburg, Russia; §School of Biomedicine, Far Eastern Federal University, Vladivostok, Russia; |Services of Pathology and ††Nephrology, Department of Medicine, University Hospital of Lausanne, Lausanne, Switzerland; and ¶Université Pierre et Marie Curie, Paris, France ABSTRACT Tight control of extracellular and intracellular inorganic phosphate (Pi) levels is crit- leaves.4 Most recently, Legati et al. have ical to most biochemical and physiologic processes. Urinary Pi is freely filtered at the shown an association between genetic kidney glomerulus and is reabsorbed in the renal tubule by the action of the apical polymorphisms in Xpr1 and primary fa- sodium-dependent phosphate transporters, NaPi-IIa/NaPi-IIc/Pit2. However, the milial brain calcification disorder.5 How- molecular identity of the protein(s) participating in the basolateral Pi efflux remains ever, the role of XPR1 in the maintenance unknown. Evidence has suggested that xenotropic and polytropic retroviral recep- of Pi homeostasis remains unknown. Here, tor 1 (XPR1) might be involved in this process. Here, we show that conditional in- we addressed this issue in mice deficient for activation of Xpr1 in the renal tubule in mice resulted in impaired renal Pi Xpr1 in the nephron. -

Alpha Actinin 4: an Intergral Component of Transcriptional
ALPHA ACTININ 4: AN INTERGRAL COMPONENT OF TRANSCRIPTIONAL PROGRAM REGULATED BY NUCLEAR HORMONE RECEPTORS By SIMRAN KHURANA Submitted in partial fulfillment of the requirements for the degree of doctor of philosophy Thesis Advisor: Dr. Hung-Ying Kao Department of Biochemistry CASE WESTERN RESERVE UNIVERSITY August, 2011 CASE WESTERN RESERVE UNIVERSITY SCHOOL OF GRADUATE STUDIES We hereby approve the thesis/dissertation of SIMRAN KHURANA ______________________________________________________ PhD candidate for the ________________________________degree *. Dr. David Samols (signed)_______________________________________________ (chair of the committee) Dr. Hung-Ying Kao ________________________________________________ Dr. Edward Stavnezer ________________________________________________ Dr. Leslie Bruggeman ________________________________________________ Dr. Colleen Croniger ________________________________________________ ________________________________________________ May 2011 (date) _______________________ *We also certify that written approval has been obtained for any proprietary material contained therein. TABLE OF CONTENTS LIST OF TABLES vii LIST OF FIGURES viii ACKNOWLEDEMENTS xii LIST OF ABBREVIATIONS xiii ABSTRACT 1 CHAPTER 1: INTRODUCTION Family of Nuclear Receptors 3 Mechanism of transcriptional regulation by co-repressors and co-activators 8 Importance of LXXLL motif of co-activators in NR mediated transcription 12 Cyclic recruitment of co-regulators on the target promoters 15 Actin and actin related proteins (ABPs) in transcription -

Genetic Background Effects of Keratin 8 and 18 in a DDC-Induced Hepatotoxicity and Mallory-Denk Body Formation Mouse Model
Laboratory Investigation (2012) 92, 857–867 & 2012 USCAP, Inc All rights reserved 0023-6837/12 $32.00 Genetic background effects of keratin 8 and 18 in a DDC-induced hepatotoxicity and Mallory-Denk body formation mouse model Johannes Haybaeck1, Cornelia Stumptner1, Andrea Thueringer1, Thomas Kolbe2, Thomas M Magin3, Michael Hesse4, Peter Fickert5, Oleksiy Tsybrovskyy1, Heimo Mu¨ller1, Michael Trauner5,6, Kurt Zatloukal1 and Helmut Denk1 Keratin 8 (K8) and keratin 18 (K18) form the major hepatocyte cytoskeleton. We investigated the impact of genetic loss of either K8 or K18 on liver homeostasis under toxic stress with the hypothesis that K8 and K18 exert different functions. krt8À/À and krt18À/À mice crossed into the same 129-ola genetic background were treated by acute and chronic ad- ministration of 3,5-diethoxy-carbonyl-1,4-dihydrocollidine (DDC). In acutely DDC-intoxicated mice, macrovesicular steatosis was more pronounced in krt8À/À and krt18À/À compared with wild-type (wt) animals. Mallory-Denk bodies (MDBs) appeared in krt18À/À mice already at an early stage of intoxication in contrast to krt8À/À mice that did not display MDB formation when fed with DDC. Keratin-deficient mice displayed significantly lower numbers of apoptotic hepatocytes than wt animals. krt8À/À, krt18À/À and control mice displayed comparable cell proliferation rates. Chronically DDC-intoxicated krt18À/À and wt mice showed a similarly increased degree of steatohepatitis with hepatocyte ballooning and MDB formation. In krt8À/À mice, steatosis was less, ballooning, and MDBs were absent. krt18À/À mice developed MDBs whereas krt8À/À mice on the same genetic background did not, highlighting the significance of different structural properties of keratins. -

Transcriptional Mechanisms of Resistance to Anti-PD-1 Therapy
Author Manuscript Published OnlineFirst on February 13, 2017; DOI: 10.1158/1078-0432.CCR-17-0270 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. Transcriptional mechanisms of resistance to anti-PD-1 therapy Maria L. Ascierto1, Alvin Makohon-Moore2, 11, Evan J. Lipson1, Janis M. Taube3,4, Tracee L. McMiller5, Alan E. Berger6, Jinshui Fan6, Genevieve J. Kaunitz3, Tricia R. Cottrell4, Zachary A. Kohutek7, Alexander Favorov8,10, Vladimir Makarov7,11, Nadeem Riaz7,11, Timothy A. Chan7,11, Leslie Cope8, Ralph H. Hruban4,9, Drew M. Pardoll1, Barry S. Taylor11,12,13, David B. Solit13, Christine A Iacobuzio-Donahue2,11, and Suzanne L. Topalian5 From the 1Departments of Oncology, 3Dermatology, 4Pathology, 5Surgery, 6The Lowe Family Genomics Core, 8Oncology Bioinformatics Core, and the 9 Sol Goldman Pancreatic Cancer Research Center, Johns Hopkins University School of Medicine and Sidney Kimmel Comprehensive Cancer Center, Baltimore, MD 21287; the 10Laboratory of System Biology and Computational Genetics, Vavilov Institute of General Genetics, Russian Academy of Sciences, 119991, Moscow, Russia; and 2Pathology, 7Radiation Oncology, 11Human Oncology and Pathogenesis Program, 12Department of Epidemiology and Biostatistics, and the 13Center for Molecular Oncology, Memorial Sloan Kettering Cancer Center, New York NY 10065. MLA, AM-M, EJL, and JMT contributed equally to this work Running title: Transcriptional mechanisms of resistance to anti-PD-1 Key Words: melanoma, cancer genetics, immunotherapy, anti-PD-1 Financial Support: This study was supported by the Melanoma Research Alliance (to SLT and CI-D), the Bloomberg~Kimmel Institute for Cancer Immunotherapy (to JMT, DMP, and SLT), the Barney Family Foundation (to SLT), Moving for Melanoma of Delaware (to SLT), the 1 Downloaded from clincancerres.aacrjournals.org on October 2, 2021. -

List of Genes Associated with Sudden Cardiac Death (Scdgseta) Gene
List of genes associated with sudden cardiac death (SCDgseta) mRNA expression in normal human heart Entrez_I Gene symbol Gene name Uniprot ID Uniprot name fromb D GTEx BioGPS SAGE c d e ATP-binding cassette subfamily B ABCB1 P08183 MDR1_HUMAN 5243 √ √ member 1 ATP-binding cassette subfamily C ABCC9 O60706 ABCC9_HUMAN 10060 √ √ member 9 ACE Angiotensin I–converting enzyme P12821 ACE_HUMAN 1636 √ √ ACE2 Angiotensin I–converting enzyme 2 Q9BYF1 ACE2_HUMAN 59272 √ √ Acetylcholinesterase (Cartwright ACHE P22303 ACES_HUMAN 43 √ √ blood group) ACTC1 Actin, alpha, cardiac muscle 1 P68032 ACTC_HUMAN 70 √ √ ACTN2 Actinin alpha 2 P35609 ACTN2_HUMAN 88 √ √ √ ACTN4 Actinin alpha 4 O43707 ACTN4_HUMAN 81 √ √ √ ADRA2B Adrenoceptor alpha 2B P18089 ADA2B_HUMAN 151 √ √ AGT Angiotensinogen P01019 ANGT_HUMAN 183 √ √ √ AGTR1 Angiotensin II receptor type 1 P30556 AGTR1_HUMAN 185 √ √ AGTR2 Angiotensin II receptor type 2 P50052 AGTR2_HUMAN 186 √ √ AKAP9 A-kinase anchoring protein 9 Q99996 AKAP9_HUMAN 10142 √ √ √ ANK2/ANKB/ANKYRI Ankyrin 2 Q01484 ANK2_HUMAN 287 √ √ √ N B ANKRD1 Ankyrin repeat domain 1 Q15327 ANKR1_HUMAN 27063 √ √ √ ANKRD9 Ankyrin repeat domain 9 Q96BM1 ANKR9_HUMAN 122416 √ √ ARHGAP24 Rho GTPase–activating protein 24 Q8N264 RHG24_HUMAN 83478 √ √ ATPase Na+/K+–transporting ATP1B1 P05026 AT1B1_HUMAN 481 √ √ √ subunit beta 1 ATPase sarcoplasmic/endoplasmic ATP2A2 P16615 AT2A2_HUMAN 488 √ √ √ reticulum Ca2+ transporting 2 AZIN1 Antizyme inhibitor 1 O14977 AZIN1_HUMAN 51582 √ √ √ UDP-GlcNAc: betaGal B3GNT7 beta-1,3-N-acetylglucosaminyltransfe Q8NFL0 -

A Key Genomic Signature Associated with Lymphovascular Invasion in Head and Neck Squamous Cell Carcinoma
A key genomic signature associated with lymphovascular invasion in head and neck squamous cell carcinoma Jian Zhang Aliated Cancer hospital & Institute of Guangzhou Medical University Huali Jiang Aliated Donghua Hospital of Sun Yat-sen University Tao Xie Aliated Cancer Hospital of Guangzhou Medical University Baiyao Wang Aliated Cancer Hospital of Guangzhou Medical Unversity Xiaoting Huang Aliated Cancer Hospital & Institute of Guangzhou Medical University Jie Lin Aliated Cancer Hospital & Institute of Guangzhou Medical University Anan Xu Aliated Cancer Hospital of Guangzhou Medical University Rong Li Aliated Cancer Hospital & Institute of Guangzhou Medical University Yawei Yuan ( [email protected] ) Guangzhou Medical University Aliated Cancer Hospital Research article Keywords: lymphovascular invasion, head and neck squamous cell carcinoma, hub genes, TCGA, weighted gene co-expression network analysis Posted Date: January 16th, 2020 DOI: https://doi.org/10.21203/rs.2.18349/v2 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/24 Abstract Objective: Lymphovascular invasion (LOI), a key pathological feature of head and neck squamous cell carcinoma (HNSCC), predicts poor survival. However, the associated clinical characteristics remain uncertain, and the molecular mechanisms are largely unknown. Methods: Weighted gene co-expression network analysis was performed to construct gene co-expression networks and investigate the relationship between modules and LOI clinical trait. Functional enrichment and KEGG pathway enrichment analysis were performed for differentially expressed genes using DAVID database. The protein-protein interaction network was constructed using Cytoscape software, and module analysis was performed using MCODE. Prognosis role and expression analysis was further validated by survival analysis, GEPIA analysis and HPA database. -

Molecular Pathways Associated with the Nutritional Programming of Plant-Based Diet Acceptance in Rainbow Trout Following an Early Feeding Exposure Mukundh N
Molecular pathways associated with the nutritional programming of plant-based diet acceptance in rainbow trout following an early feeding exposure Mukundh N. Balasubramanian, Stéphane Panserat, Mathilde Dupont-Nivet, Edwige Quillet, Jérôme Montfort, Aurélie Le Cam, Françoise Médale, Sadasivam J. Kaushik, Inge Geurden To cite this version: Mukundh N. Balasubramanian, Stéphane Panserat, Mathilde Dupont-Nivet, Edwige Quillet, Jérôme Montfort, et al.. Molecular pathways associated with the nutritional programming of plant-based diet acceptance in rainbow trout following an early feeding exposure. BMC Genomics, BioMed Central, 2016, 17 (1), pp.1-20. 10.1186/s12864-016-2804-1. hal-01346912 HAL Id: hal-01346912 https://hal.archives-ouvertes.fr/hal-01346912 Submitted on 19 Jul 2016 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Balasubramanian et al. BMC Genomics (2016) 17:449 DOI 10.1186/s12864-016-2804-1 RESEARCH ARTICLE Open Access Molecular pathways associated with the nutritional programming of plant-based diet acceptance in rainbow trout following an early feeding exposure Mukundh N. Balasubramanian1, Stephane Panserat1, Mathilde Dupont-Nivet2, Edwige Quillet2, Jerome Montfort3, Aurelie Le Cam3, Francoise Medale1, Sadasivam J. Kaushik1 and Inge Geurden1* Abstract Background: The achievement of sustainable feeding practices in aquaculture by reducing the reliance on wild-captured fish, via replacement of fish-based feed with plant-based feed, is impeded by the poor growth response seen in fish fed high levels of plant ingredients. -
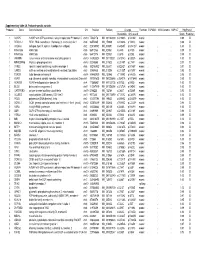
Supplementary Table 2A. Proband-Specific Variants Proband
Supplementary table 2A. Proband-specific variants Proband Gene Gene full name Chr Position RefSeqChange Function ESP6500 1000 Genome SNP137 PolyPhen2 Nucleotide Amino acid Score Prediction 1 AGAP5 ArfGAP with GTPase domain, ankyrin repeat and PH domain 5 chr10 75435178 NM_001144000 c.G1240A p.V414M exonic . 0.99 D 1 REXO1L1 REX1, RNA exonuclease 1 homolog (S. cerevisiae)-like 1 chr8 86574465 NM_172239 c.A1262G p.Y421C exonic . 0.98 D 1 COL4A3 collagen, type IV, alpha 3 (Goodpasture antigen) chr2 228169782 NM_000091 c.G4235T p.G1412V exonic . 1.00 D 1 KIAA1586 KIAA1586 chr6 56912133 NM_020931 c.C44G p.A15G exonic . 0.99 D 1 KIAA1586 KIAA1586 chr6 56912176 NM_020931 c.C87G p.D29E exonic . 0.99 D 1 JAKMIP3 Janus kinase and microtubule interacting protein 3 chr10 133955524 NM_001105521 c.G1574C p.G525A exonic . 1.00 D 1 MPHOSPH8 M-phase phosphoprotein 8 chr13 20240685 NM_017520 c.C2140T p.L714F exonic . 0.97 D 1 SYNE1 spectrin repeat containing, nuclear envelope 1 chr6 152783902 NM_033071 c.A2242T p.N748Y exonic . 0.97 D 1 CAND2 cullin-associated and neddylation-dissociated 2 (putative) chr3 12858835 NM_012298 c.C2125T p.H709Y exonic . 0.95 D 1 TDRD9 tudor domain containing 9 chr14 104460909 NM_153046 c.T1289G p.V430G exonic . 0.96 D 2 ASPM asp (abnormal spindle) homolog, microcephaly associated (Drosophila)chr1 197091625 NM_001206846 c.G3491A p.R1164H exonic . 0.99 D 2 ADAM29 ADAM metallopeptidase domain 29 chr4 175896951 NM_001130705 c.A275G p.D92G exonic . 1.00 D 2 BCO2 beta-carotene oxygenase 2 chr11 112087019 NM_001256398 c.G1373A p.G458E exonic . -

Structural and Biochemical Changes Underlying a Keratoderma-Like Phenotype in Mice Lacking Suprabasal AP1 Transcription Factor Function
Citation: Cell Death and Disease (2015) 6, e1647; doi:10.1038/cddis.2015.21 OPEN & 2015 Macmillan Publishers Limited All rights reserved 2041-4889/15 www.nature.com/cddis Structural and biochemical changes underlying a keratoderma-like phenotype in mice lacking suprabasal AP1 transcription factor function EA Rorke*,1, G Adhikary2, CA Young2, RH Rice3, PM Elias4, D Crumrine4, J Meyer4, M Blumenberg5 and RL Eckert2,6,7,8 Epidermal keratinocyte differentiation on the body surface is a carefully choreographed process that leads to assembly of a barrier that is essential for life. Perturbation of keratinocyte differentiation leads to disease. Activator protein 1 (AP1) transcription factors are key controllers of this process. We have shown that inhibiting AP1 transcription factor activity in the suprabasal murine epidermis, by expression of dominant-negative c-jun (TAM67), produces a phenotype type that resembles human keratoderma. However, little is understood regarding the structural and molecular changes that drive this phenotype. In the present study we show that TAM67-positive epidermis displays altered cornified envelope, filaggrin-type keratohyalin granule, keratin filament, desmosome formation and lamellar body secretion leading to reduced barrier integrity. To understand the molecular changes underlying this process, we performed proteomic and RNA array analysis. Proteomic study of the corneocyte cross-linked proteome reveals a reduction in incorporation of cutaneous keratins, filaggrin, filaggrin2, late cornified envelope precursor proteins, hair keratins and hair keratin-associated proteins. This is coupled with increased incorporation of desmosome linker, small proline-rich, S100, transglutaminase and inflammation-associated proteins. Incorporation of most cutaneous keratins (Krt1, Krt5 and Krt10) is reduced, but incorporation of hyperproliferation-associated epidermal keratins (Krt6a, Krt6b and Krt16) is increased.