Characterization of the Uncommon Enzymes from (2004)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
A Deep Learning Approach to Pattern Recognition for Short DNA Sequences”
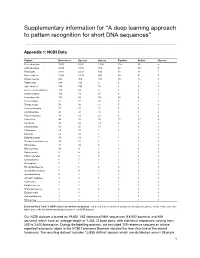
Supplementary information for “A deep learning approach to pattern recognition for short DNA sequences” Appendix 1: NCBI Data Phylum References Species Genera Families Orders Classes Proteobacteria 7,053 5,061 1,106 158 55 9 Actinobacteria 4,768 3,313 383 68 29 6 Firmicutes 3,814 2,531 499 56 13 7 Bacteroidetes 1,934 1,525 360 39 8 7 Euryarchaeota 834 450 100 28 13 8 Tenericutes 266 195 8 5 4 1 Spirochaetes 146 100 16 6 4 1 Deinococcus-Thermus 118 99 9 3 2 1 Crenarchaeota 113 61 27 8 5 1 Cyanobacteria 113 88 59 30 8 2 Fusobacteria 75 37 10 2 1 1 Thermotogae 70 48 13 5 4 1 Verrucomicrobia 57 53 22 7 4 3 Acidobacteria 45 41 19 5 5 4 Planctomycetes 44 31 23 5 3 2 Chloroflexi 44 35 26 15 12 8 Aquificae 43 32 14 4 2 1 Synergistetes 31 25 15 1 1 1 Chlamydiae 28 18 7 5 2 1 Chlorobi 21 16 5 1 1 1 Deferribacteres 15 11 7 1 1 1 Thermodesulfobacteria 14 12 5 1 1 1 Nitrospirae 11 10 3 1 1 1 Fibrobacteres 10 4 3 3 3 3 Balneolaeota 9 9 4 1 1 1 Chrysiogenetes 6 4 3 1 1 1 Lentisphaerae 6 5 3 3 3 2 Dictyoglomi 5 2 1 1 1 1 Rhodothermaeota 5 5 3 2 1 1 Gemmatimonadetes 5 4 3 2 2 2 Ignavibacteriae 4 2 2 2 1 1 Armatimonadetes 4 3 3 3 3 3 Caldiserica 3 1 1 1 1 1 Calditrichaeota 2 2 1 1 1 1 Thaumarchaeota 2 2 2 2 2 2 Elusimicrobia 2 1 1 1 1 1 Kiritimatiellaeota 1 1 1 1 1 1 Nitrospinae 1 1 1 1 1 1 Extended Data Table 1: NCBI dataset breakdown by phylum. -

Development of Bacterial Communities in Biological Soil Crusts Along
1 Development of bacterial communities in biological soil crusts along 2 a revegetation chronosequence in the Tengger Desert, northwest 3 China 4 5 Author names and affiliations: 6 Lichao Liu1, Yubing Liu1, 2 *, Peng Zhang1, Guang Song1, Rong Hui1, Zengru Wang1, Jin Wang1, 2 7 1Shapotou Desert Research & Experiment Station, Northwest Institute of Eco-Environment and Resources, Chinese 8 Academy of Sciences, Lanzhou, 730000, China 9 2Key Laboratory of Stress Physiology and Ecology in Cold and Arid Regions of Gansu Province, Northwest Institute 10 of Eco–Environment and Resources, Chinese Academy of Sciences, Lanzhou 730000, China 11 12 * Corresponding author: Yubing Liu 13 Address: Donggang West Road 320, Lanzhou 730000, P. R. China. 14 Tel: +86 0931 4967202. 15 E-mail address: [email protected] 16 17 Abstract. Knowledge of structure and function of microbial communities in different 18 successional stages of biological soil crusts (BSCs) is still scarce for desert areas. In this study, 19 Illumina MiSeq sequencing was used to assess the composition changes of bacterial communities 20 in different ages of BSCs in the revegetation of Shapotou in the Tengger Desert. The most dominant 21 phyla of bacterial communities shifted with the changed types of BSCs in the successional stages, 22 from Firmicutes in mobile sand and physical crusts to Actinobacteria and Proteobacteria in BSCs, 23 and the most dominant genera shifted from Bacillus, Enterococcus and Lactococcus to 24 RB41_norank and JG34-KF-361_norank. Alpha diversity and quantitative real-time PCR analysis 25 indicated that bacteria richness and abundance reached their highest levels after 15 years of BSC 26 development. -

7. References
University of Akureyri Department of Natural Resource Science 7. References Ammann, E. C., Reed, L. L., & Durichek, J. J. (1968). Gas consumptions and Growth Rate of Hydrogenomonas eutropha in Continuous Culture. Applied Microbiology , 16, (6), 822-826. Aguiar, P., Beveridge, T. J., & Reysenbach, A.-L. (2004). Sulfurihydrogenibium azorense, sp. nov., a thermophilic hydrogen oxidizing microaerophile from terrestrial hot springs in the Azores. International Journal of Systematic and Evolutionary Microbiology , 54, 33-39. Altschul, S., Gish, W., Miller, W., Myers, E., & Lipman, D. (1990). "Basic local alignment search tool.". J. Mol. Biol. , 215:403-410. Amend, J., & Shock, E. (2001). Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and Bacteria. FEMS Microbiology Reviews , 25, 175-243. Aragno, M. (1978). Enrichment, isolation and preliminary characterization of a thermophilic, endospore-forming hydrogen bacterium. FEMS Micobiol. Lett. , 3: 13-15. Aragno, M. (1992). The Thermophilic, Aerobic, Hydrogen-Oxidizing (Knallgas) Bacteria. In A. Balows, H. Trüper, M. Dworkin, W. Harder, & K. Schleifer, The Prokaryotes, a handbook on biology of bacteria. 2nd ed. vol. 4 (pp. 3917-3933.). New York: Springer Verlag. Aragno, M., & Schlegel, H. G. (1992). The mesophilic Hydrogen-Oxidizing (Knallgas) Bacteria. In A. Balows, H. Truper, M. Dworkin, W. Harder, & K.-H. Schleifer, The Prokaryotes 2nd. ed. (pp. 344-384). New York: Springer. Ármannson, H. (2002, May 30-31). Erindi á ráðstefnu um málefni veitufyrirtækja . Grænt bókhald í jarðhita- samanburður á útblæstri við aðra orkugjafa . Akureyri, Iceland: Samorka. Bae, S., Kwak, K., Kim, S., Chung, S., & Igarashi, Y. (2001). Isolation and Characterization of CO2-Fixing Hydrogen -Oxidizing Marine 109 University of Akureyri Department of Natural Resource Science Bacteria. -

And Thermo-Adaptation in Hyperthermophilic Archaea: Identification of Compatible Solutes, Accumulation Profiles, and Biosynthetic Routes in Archaeoglobus Spp
Universidade Nova de Lisboa Osmo- andInstituto thermo de Tecnologia-adaptation Química e Biológica in hyperthermophilic Archaea: Subtitle Subtitle Luís Pedro Gafeira Gonçalves Osmo- and thermo-adaptation in hyperthermophilic Archaea: identification of compatible solutes, accumulation profiles, and biosynthetic routes in Archaeoglobus spp. OH OH OH CDP c c c - CMP O O - PPi O3P P CTP O O O OH OH OH OH OH OH O- C C C O P O O P i Dissertation presented to obtain the Ph.D degree in BiochemistryO O- Instituto de Tecnologia Química e Biológica | Universidade Nova de LisboaP OH O O OH OH OH Oeiras, Luís Pedro Gafeira Gonçalves January, 2008 2008 Universidade Nova de Lisboa Instituto de Tecnologia Química e Biológica Osmo- and thermo-adaptation in hyperthermophilic Archaea: identification of compatible solutes, accumulation profiles, and biosynthetic routes in Archaeoglobus spp. This dissertation was presented to obtain a Ph. D. degree in Biochemistry at the Instituto de Tecnologia Química e Biológica, Universidade Nova de Lisboa. By Luís Pedro Gafeira Gonçalves Supervised by Prof. Dr. Helena Santos Oeiras, January, 2008 Apoio financeiro da Fundação para a Ciência e Tecnologia (POCI 2010 – Formação Avançada para a Ciência – Medida IV.3) e FSE no âmbito do Quadro Comunitário de apoio, Bolsa de Doutoramento com a referência SFRH / BD / 5076 / 2001. ii ACKNOWNLEDGMENTS The work presented in this thesis, would not have been possible without the help, in terms of time and knowledge, of many people, to whom I am extremely grateful. Firstly and mostly, I need to thank my supervisor, Prof. Helena Santos, for her way of thinking science, her knowledge, her rigorous criticism, and her commitment to science. -

Levels of Firmicutes, Actinobacteria Phyla and Lactobacillaceae
agriculture Article Levels of Firmicutes, Actinobacteria Phyla and Lactobacillaceae Family on the Skin Surface of Broiler Chickens (Ross 308) Depending on the Nutritional Supplement and the Housing Conditions Paulina Cholewi ´nska 1,* , Marta Michalak 2, Konrad Wojnarowski 1 , Szymon Skowera 1, Jakub Smoli ´nski 1 and Katarzyna Czyz˙ 1 1 Institute of Animal Breeding, Wroclaw University of Environmental and Life Sciences, 51-630 Wroclaw, Poland; [email protected] (K.W.); [email protected] (S.S.); [email protected] (J.S.); [email protected] (K.C.) 2 Department of Animal Nutrition and Feed Management, Wroclaw University of Environmental and Life Sciences, 51-630 Wroclaw, Poland; [email protected] * Correspondence: [email protected] Abstract: The microbiome of animals, both in the digestive tract and in the skin, plays an important role in protecting the host. The skin is one of the largest surface organs for animals; therefore, the destabilization of the microbiota on its surface can increase the risk of diseases that may adversely af- fect animals’ health and production rates, including poultry. The aim of this study was to evaluate the Citation: Cholewi´nska,P.; Michalak, effect of nutritional supplementation in the form of fermented rapeseed meal and housing conditions M.; Wojnarowski, K.; Skowera, S.; on the level of selected bacteria phyla (Firmicutes, Actinobacteria, and family Lactobacillaceae). The Smoli´nski,J.; Czyz,˙ K. Levels of study was performed on 30 specimens of broiler chickens (Ross 308), individually kept in metabolic Firmicutes, Actinobacteria Phyla and cages for 36 days. They were divided into 5 groups depending on the feed received. -

Molecular Evolution of the Oxygen-Binding Hemerythrin Domain
RESEARCH ARTICLE Molecular Evolution of the Oxygen-Binding Hemerythrin Domain Claudia Alvarez-Carreño1, Arturo Becerra1, Antonio Lazcano1,2* 1 Facultad de Ciencias, Universidad Nacional Autónoma de México, Apdo. Postal 70–407, Cd. Universitaria, 04510, Mexico City, Mexico, 2 Miembro de El Colegio Nacional, Ciudad de México, México * [email protected] a11111 Abstract Background The evolution of oxygenic photosynthesis during Precambrian times entailed the diversifica- tion of strategies minimizing reactive oxygen species-associated damage. Four families of OPEN ACCESS oxygen-carrier proteins (hemoglobin, hemerythrin and the two non-homologous families of Citation: Alvarez-Carreño C, Becerra A, Lazcano A arthropodan and molluscan hemocyanins) are known to have evolved independently the (2016) Molecular Evolution of the Oxygen-Binding Hemerythrin Domain. PLoS ONE 11(6): e0157904. capacity to bind oxygen reversibly, providing cells with strategies to cope with the evolution- doi:10.1371/journal.pone.0157904 ary pressure of oxygen accumulation. Oxygen-binding hemerythrin was first studied in Editor: Nikolas Nikolaidis, California State University marine invertebrates but further research has made it clear that it is present in the three Fullerton, UNITED STATES domains of life, strongly suggesting that its origin predated the emergence of eukaryotes. Received: April 5, 2016 Accepted: June 7, 2016 Results Published: June 23, 2016 Oxygen-binding hemerythrins are a monophyletic sub-group of the hemerythrin/HHE (histi- dine, histidine, glutamic acid) cation-binding domain. Oxygen-binding hemerythrin homo- Copyright: © 2016 Alvarez-Carreño et al. This is an open access article distributed under the terms of the logs were unambiguously identified in 367/2236 bacterial, 21/150 archaeal and 4/135 Creative Commons Attribution License, which permits eukaryotic genomes. -

A Study of the Microorganisms from Grass Silage I
A Study of the Microorganisms from Grass Silage I. The Cocci C. WV. LANGSTON AND CECELIA BOUMA Dairy Cattle Research Branch, Animal Husbandry Research Division, Agriculture Research Service, 1griculture Research Center, Beltsville, Marlyland Received for ptiblication November 16, 1959 In a natural silage fermentation, the mass is acidified their taxonomical relationship. The data presented are by lactic and acetic acid forming bacteria that ferment the results of detailed colonial, morphological, and sugars in the plant material. When forage is ensiled the physiological studies on the cocci important in the plant cells continue to respire for a time, using up the silage fermentation. oxygen and giving off CO2 and heat. As conditions be- come favorable, acid producing bacteria increase MIATERI.ALS ANDV IETHODS rapidly and, at the end of 3 or 4 days, each gram of Preparationi of silages and techniques used in iso- silage will contain several hundred million bacteria. lating and grouping the lactic acid bacteria from the These organisms produce acid until the sugar is ex- silages have been outlined in an earlier publication hausted or until the pH becomes unfavorable for further (Langston et al., 1958). growth. This phase of w-ork includes detailed studies on repre- A recent study (Langston et al., 1958) on the micro- sentative strains of lactic acid bacteria obtained from organisms in orchard grass and alfalfa silages showed 30 silages. The strains were picked from highest dilution that the total numbers of acid producing bacteria had roll tubes (Trypticase)l and plates (Rogosa et al., 1951). little bearing on the final quality. -

Endocarditis Caused by Leuconostoc Lactis in an Infant. Case Report Endocarditis Por Leuconostoc Lactis En Un Lactante
467 Rev. Fac. Med. 2020 Vol. 68 No. 3: 467-70 CASE REPORT DOI: http://dx.doi.org/10.15446/revfacmed.v68n3.77425 Received: 22/01/2019 Accepted: 14/04/2019 Revista de la Facultad de Medicina Endocarditis caused by Leuconostoc lactis in an infant. Case report Endocarditis por Leuconostoc lactis en un lactante. Reporte de caso Edgar Alberto Sarmiento-Ortiz1, Oskar Andrey Oliveros-Andrade1,2, Juan Pablo Rojas-Hernández1,2,3,4 1 Universidad Libre Cali Campus - Faculty of Health Sciences - Department of Pediatrics - Pediatrics Research Group - Santiago de Cali - Colombia. 2 Universidad Libre Cali Campus - Faculty of Health Sciences - Department of Pediatrics - Santiago de Cali - Colombia. 3 Fundación Clínica Infantil Club Noel - Department of Infectious Diseases - Santiago de Cali - Colombia. 4 Universidad del Valle - Faculty of Health - Santiago de Cali - Colombia. Corresponding auhtor: Oskar Andrey Oliveros-Andrade. Departamento de Pediatría, Facultad de Ciencias de la Salud, Universidad Libre Seccional Cali. Carrera 109 No. 22-00, bloque: 5, oficina de Posgrados Clínicos de la Facultad de Ciencias de la Salud. Telephone number: +57 2 5240007, ext.: 2543. Santiago de Cali. Colombia. Email: [email protected]. Abstract Introduction: Infections caused by Leuconostoc lactis are rare and are associated with Sarmiento-Ortiz EA, Oliveros-Andra- multiple risk factors. According to the literature reviewed, there are no reported cases of de OA, Rojas-Hernández JP. Endocar- ditis caused by Leuconostoc Lactis in endocarditis caused by this microorganism in the pediatric population. an infant. Case report. Rev. Fac. Med. Case presentation: An infant with short bowel syndrome was taken by his parents to the 2020;68(3):467-70. -

Across the Tree of Life, Radiation Resistance Is Governed By
Across the tree of life, radiation resistance is PNAS PLUS + governed by antioxidant Mn2 , gauged by paramagnetic resonance Ajay Sharmaa,1, Elena K. Gaidamakovab,c,1, Olga Grichenkob,c, Vera Y. Matrosovab,c, Veronika Hoekea, Polina Klimenkovab,c, Isabel H. Conzeb,d, Robert P. Volpeb,c, Rok Tkavcb,c, Cene Gostincarˇ e, Nina Gunde-Cimermane, Jocelyne DiRuggierof, Igor Shuryakg, Andrew Ozarowskih, Brian M. Hoffmana,i,2, and Michael J. Dalyb,2 aDepartment of Chemistry, Northwestern University, Evanston, IL 60208; bDepartment of Pathology, Uniformed Services University of the Health Sciences, Bethesda, MD 20814; cHenry M. Jackson Foundation for the Advancement of Military Medicine, Bethesda, MD 20817; dDepartment of Biology, University of Bielefeld, Bielefeld, 33615, Germany; eDepartment of Biology, Biotechnical Faculty, University of Ljubljana, Ljubljana, SI-1000, Slovenia; fDepartment of Biology, Johns Hopkins University, Baltimore, MD 21218; gCenter for Radiological Research, Columbia University, New York, NY 10032; hNational High Magnetic Field Laboratory, Florida State University, Tallahassee, FL 32306; and iDepartment of Molecular Biosciences, Northwestern University, Evanston, IL 60208 Contributed by Brian M. Hoffman, September 15, 2017 (sent for review August 1, 2017; reviewed by Valeria Cizewski Culotta and Stefan Stoll) Despite concerted functional genomic efforts to understand the agents of cellular damage. In particular, irradiated cells rapidly form •− complex phenotype of ionizing radiation (IR) resistance, a genome superoxide (O2 ) ions by radiolytic reduction of both atmospheric sequence cannot predict whether a cell is IR-resistant or not. Instead, O2 and O2 released through the intracellular decomposition of IR- we report that absorption-display electron paramagnetic resonance generated H2O2 ascatalyzedbybothenzymaticandnonenzymatic (EPR) spectroscopy of nonirradiated cells is highly diagnostic of IR •− metal ions. -

Comparative Analyses of Whole-Genome Protein Sequences
www.nature.com/scientificreports OPEN Comparative analyses of whole- genome protein sequences from multiple organisms Received: 7 June 2017 Makio Yokono 1,2, Soichirou Satoh3 & Ayumi Tanaka1 Accepted: 16 April 2018 Phylogenies based on entire genomes are a powerful tool for reconstructing the Tree of Life. Several Published: xx xx xxxx methods have been proposed, most of which employ an alignment-free strategy. Average sequence similarity methods are diferent than most other whole-genome methods, because they are based on local alignments. However, previous average similarity methods fail to reconstruct a correct phylogeny when compared against other whole-genome trees. In this study, we developed a novel average sequence similarity method. Our method correctly reconstructs the phylogenetic tree of in silico evolved E. coli proteomes. We applied the method to reconstruct a whole-proteome phylogeny of 1,087 species from all three domains of life, Bacteria, Archaea, and Eucarya. Our tree was automatically reconstructed without any human decisions, such as the selection of organisms. The tree exhibits a concentric circle-like structure, indicating that all the organisms have similar total branch lengths from their common ancestor. Branching patterns of the members of each phylum of Bacteria and Archaea are largely consistent with previous reports. The topologies are largely consistent with those reconstructed by other methods. These results strongly suggest that this approach has sufcient taxonomic resolution and reliability to infer phylogeny, from phylum to strain, of a wide range of organisms. Te reconstruction of phylogenetic trees is a powerful tool for understanding organismal evolutionary processes. Molecular phylogenetic analysis using ribosomal RNA (rRNA) clarifed the phylogenetic relationship of the three domains, bacterial, archaeal, and eukaryotic1. -

Download PDF (Inglês)
b r a z i l i a n j o u r n a l o f m i c r o b i o l o g y 4 9 (2 0 1 8) 248–257 ht tp://www.bjmicrobiol.com.br/ Environmental Microbiology Analysis of bacterial communities and characterization of antimicrobial strains from cave microbiota Muhammad Yasir King Abdulaziz University, King Fahd Medical Research Center, Special Infectious Agents Unit, Jeddah, Saudi Arabia a r a t b i c l e i n f o s t r a c t Article history: In this study for the first-time microbial communities in the caves located in the mountain Received 12 October 2016 range of Hindu Kush were evaluated. The samples were analyzed using culture-independent Accepted 15 August 2017 (16S rRNA gene amplicon sequencing) and culture-dependent methods. The amplicon Available online 18 October 2017 sequencing results revealed a broad taxonomic diversity, including 21 phyla and 20 candidate phyla. Proteobacteria were dominant in both caves, followed by Bacteroidetes, Acti- Associate Editor: John McCulloch nobacteria, Firmicutes, Verrucomicrobia, Planctomycetes, and the archaeal phylum Euryarchaeota. Representative operational taxonomic units from Koat Maqbari Ghaar and Smasse-Rawo Keywords: Ghaar were grouped into 235 and 445 different genera, respectively. Comparative analysis Caves of the cultured bacterial isolates revealed distinct bacterial taxonomic profiles in the stud- 16S ribosomal RNA ied caves dominated by Proteobacteria in Koat Maqbari Ghaar and Firmicutes in Smasse-Rawo Microbiota Ghaar. Majority of those isolates were associated with the genera Pseudomonas and Bacillus. Antimicrobial Thirty strains among the identified isolates from both caves showed antimicrobial activ- Sediments ity. -

NOC04013 Application.Pdf(PDF, 453
ER-AN-02N 10/02 Application for approval to import into FORM 2N containment any new organism that is not genetically modified, under Section 40 of the Page 1 Hazardous Substances and New Organisms Act 1996 FORM NO2N Application for approval to IMPORT INTO CONTAINMENT ANY NEW ORGANISM THAT IS NOT GENETICALLY MODIFIED under section 40 of the Hazardous Substances and New Organisms Act 1996 Application Title: Importation of sediment/water samples collected from hydrothermal seamounts outside New Zealand Territorial waters containing Extremophilic microorganisms Applicant Organisation: Institute of Geological & Nuclear Sciences ERMA Office use only Application Code: Formally received:____/____/____ ERMA NZ Contact: Initial Fee Paid: $ Application Status: ER-AN-02N 10/02 Application for approval to import into FORM 2N containment any new organism that is not genetically modified, under Section 40 of the Page 2 Hazardous Substances and New Organisms Act 1996 IMPORTANT 1. An associated User Guide is available for this form. You should read the User Guide before completing this form. If you need further guidance in completing this form please contact ERMA New Zealand. 2. This application form covers importation into containment of any new organism that is not genetically modified, under section 40 of the Act. 3. If you are making an application to import into containment a genetically modified organism you should complete Form NO2G, instead of this form (Form NO2N). 4. This form, together with form NO2G, replaces all previous versions of Form 2. Older versions should not now be used. You should periodically check with ERMA New Zealand or on the ERMA New Zealand web site for new versions of this form.