Molecular Evolution of the Oxygen-Binding Hemerythrin Domain
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genomics 98 (2011) 370–375
Genomics 98 (2011) 370–375 Contents lists available at ScienceDirect Genomics journal homepage: www.elsevier.com/locate/ygeno Whole-genome comparison clarifies close phylogenetic relationships between the phyla Dictyoglomi and Thermotogae Hiromi Nishida a,⁎, Teruhiko Beppu b, Kenji Ueda b a Agricultural Bioinformatics Research Unit, Graduate School of Agricultural and Life Sciences, University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo 113-8657, Japan b Life Science Research Center, College of Bioresource Sciences, Nihon University, Fujisawa, Japan article info abstract Article history: The anaerobic thermophilic bacterial genus Dictyoglomus is characterized by the ability to produce useful Received 2 June 2011 enzymes such as amylase, mannanase, and xylanase. Despite the significance, the phylogenetic position of Accepted 1 August 2011 Dictyoglomus has not yet been clarified, since it exhibits ambiguous phylogenetic positions in a single gene Available online 7 August 2011 sequence comparison-based analysis. The number of substitutions at the diverging point of Dictyoglomus is insufficient to show the relationships in a single gene comparison-based analysis. Hence, we studied its Keywords: evolutionary trait based on whole-genome comparison. Both gene content and orthologous protein sequence Whole-genome comparison Dictyoglomus comparisons indicated that Dictyoglomus is most closely related to the phylum Thermotogae and it forms a Bacterial systematics monophyletic group with Coprothermobacter proteolyticus (a constituent of the phylum Firmicutes) and Coprothermobacter proteolyticus Thermotogae. Our findings indicate that C. proteolyticus does not belong to the phylum Firmicutes and that the Thermotogae phylum Dictyoglomi is not closely related to either the phylum Firmicutes or Synergistetes but to the phylum Thermotogae. © 2011 Elsevier Inc. -
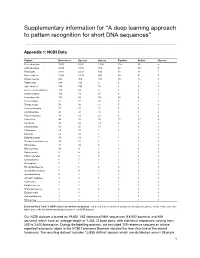
A Deep Learning Approach to Pattern Recognition for Short DNA Sequences”
Supplementary information for “A deep learning approach to pattern recognition for short DNA sequences” Appendix 1: NCBI Data Phylum References Species Genera Families Orders Classes Proteobacteria 7,053 5,061 1,106 158 55 9 Actinobacteria 4,768 3,313 383 68 29 6 Firmicutes 3,814 2,531 499 56 13 7 Bacteroidetes 1,934 1,525 360 39 8 7 Euryarchaeota 834 450 100 28 13 8 Tenericutes 266 195 8 5 4 1 Spirochaetes 146 100 16 6 4 1 Deinococcus-Thermus 118 99 9 3 2 1 Crenarchaeota 113 61 27 8 5 1 Cyanobacteria 113 88 59 30 8 2 Fusobacteria 75 37 10 2 1 1 Thermotogae 70 48 13 5 4 1 Verrucomicrobia 57 53 22 7 4 3 Acidobacteria 45 41 19 5 5 4 Planctomycetes 44 31 23 5 3 2 Chloroflexi 44 35 26 15 12 8 Aquificae 43 32 14 4 2 1 Synergistetes 31 25 15 1 1 1 Chlamydiae 28 18 7 5 2 1 Chlorobi 21 16 5 1 1 1 Deferribacteres 15 11 7 1 1 1 Thermodesulfobacteria 14 12 5 1 1 1 Nitrospirae 11 10 3 1 1 1 Fibrobacteres 10 4 3 3 3 3 Balneolaeota 9 9 4 1 1 1 Chrysiogenetes 6 4 3 1 1 1 Lentisphaerae 6 5 3 3 3 2 Dictyoglomi 5 2 1 1 1 1 Rhodothermaeota 5 5 3 2 1 1 Gemmatimonadetes 5 4 3 2 2 2 Ignavibacteriae 4 2 2 2 1 1 Armatimonadetes 4 3 3 3 3 3 Caldiserica 3 1 1 1 1 1 Calditrichaeota 2 2 1 1 1 1 Thaumarchaeota 2 2 2 2 2 2 Elusimicrobia 2 1 1 1 1 1 Kiritimatiellaeota 1 1 1 1 1 1 Nitrospinae 1 1 1 1 1 1 Extended Data Table 1: NCBI dataset breakdown by phylum. -

Sporulation Evolution and Specialization in Bacillus
bioRxiv preprint doi: https://doi.org/10.1101/473793; this version posted March 11, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Research article From root to tips: sporulation evolution and specialization in Bacillus subtilis and the intestinal pathogen Clostridioides difficile Paula Ramos-Silva1*, Mónica Serrano2, Adriano O. Henriques2 1Instituto Gulbenkian de Ciência, Oeiras, Portugal 2Instituto de Tecnologia Química e Biológica, Universidade Nova de Lisboa, Oeiras, Portugal *Corresponding author: Present address: Naturalis Biodiversity Center, Marine Biodiversity, Leiden, The Netherlands Phone: 0031 717519283 Email: [email protected] (Paula Ramos-Silva) Running title: Sporulation from root to tips Keywords: sporulation, bacterial genome evolution, horizontal gene transfer, taxon- specific genes, Bacillus subtilis, Clostridioides difficile 1 bioRxiv preprint doi: https://doi.org/10.1101/473793; this version posted March 11, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Abstract Bacteria of the Firmicutes phylum are able to enter a developmental pathway that culminates with the formation of a highly resistant, dormant spore. Spores allow environmental persistence, dissemination and for pathogens, are infection vehicles. In both the model Bacillus subtilis, an aerobic species, and in the intestinal pathogen Clostridioides difficile, an obligate anaerobe, sporulation mobilizes hundreds of genes. -

Evolution Génomique Chez Les Bactéries Du Super Phylum Planctomycetes-Verrucomicrobiae-Chlamydia
AIX-MARSEILLE UNIVERSITE FACULTE DE MEDECINE DE MARSEILLE ECOLE DOCTORALE : SCIENCE DE LA VIE ET DE LA SANTE THESE Présentée et publiquement soutenue devant LA FACULTE DE MEDECINE DE MARSEILLE Le 15 janvier 2016 Par Mme Sandrine PINOS Née à Saint-Gaudens le 09 octobre 1989 TITRE DE LA THESE: Evolution génomique chez les bactéries du super phylum Planctomycetes-Verrucomicrobiae-Chlamydia Pour obtenir le grade de DOCTORAT d'AIX-MARSEILLE UNIVERSITE Spécialité : Génomique et Bioinformatique Membres du jury de la Thèse: Pr Didier RAOULT .................................................................................Directeur de thèse Dr Pierre PONTAROTTI ....................................................................Co-directeur de thèse Pr Gilbert GREUB .............................................................................................Rapporteur Dr Pascal SIMONET............................................................................................Rapporteur Laboratoires d’accueil Unité de Recherche sur les Maladies Infectieuses et Tropicales Emergentes – UMR CNRS 6236, IRD 198 I2M - UMR CNRS 7373 - EBM 1 Avant propos Le format de présentation de cette thèse correspond à une recommandation de la spécialité Maladies Infectieuses et Microbiologie, à l’intérieur du Master de Sciences de la Vie et de la Santé qui dépend de l’Ecole Doctorale des Sciences de la Vie de Marseille. Le candidat est amené à respecter des règles qui lui sont imposées et qui comportent un format de thèse utilisé dans le Nord de l’Europe permettant un meilleur rangement que les thèses traditionnelles. Par ailleurs, la partie introduction et bibliographie est remplacée par une revue envoyée dans un journal afin de permettre une évaluation extérieure de la qualité de la revue et de permettre à l’étudiant de le commencer le plus tôt possible une bibliographie exhaustive sur le domaine de cette thèse. Par ailleurs, la thèse est présentée sur article publié, accepté ou soumis associé d’un bref commentaire donnant le sens général du travail. -

Supplemental Material S1.Pdf
Phylogeny of Selenophosphate synthetases (SPS) Supplementary Material S1 ! SelD in prokaryotes! ! ! SelD gene finding in sequenced prokaryotes! We downloaded a total of 8263 prokaryotic genomes from NCBI (see Supplementary Material S7). We scanned them with the program selenoprofiles (Mariotti 2010, http:// big.crg.cat/services/selenoprofiles) using two SPS-family profiles, one prokaryotic (seld) and one mixed eukaryotic-prokaryotic (SPS). Selenoprofiles removes overlapping predictions from different profiles, keeping only the prediction from the profile that seems closer to the candidate sequence. As expected, the great majority of output predictions in prokaryotic genomes were from the seld profile. We will refer to the prokaryotic SPS/SelD !genes as SelD, following the most common nomenclature in literature.! To be able to inspect results by hand, and also to focus on good-quality genomes, we considered a reduced set of species. We took the prok_reference_genomes.txt list from ftp://ftp.ncbi.nlm.nih.gov/genomes/GENOME_REPORTS/, which NCBI claims to be a "small curated subset of really good and scientifically important prokaryotic genomes". We named this the prokaryotic reference set (223 species - see Supplementary Material S8). We manually curated most of the analysis in this set, while we kept automatized the !analysis on the full set.! We detected SelD proteins in 58 genomes (26.0%) in the prokaryotic reference set (figure 1 in main paper), which become 2805 (33.9%) when considering the prokaryotic full set (figure SM1.1). The difference in proportion between the two sets is due largely to the presence of genomes of very close strains in the full set, which we consider redundant. -

7. References
University of Akureyri Department of Natural Resource Science 7. References Ammann, E. C., Reed, L. L., & Durichek, J. J. (1968). Gas consumptions and Growth Rate of Hydrogenomonas eutropha in Continuous Culture. Applied Microbiology , 16, (6), 822-826. Aguiar, P., Beveridge, T. J., & Reysenbach, A.-L. (2004). Sulfurihydrogenibium azorense, sp. nov., a thermophilic hydrogen oxidizing microaerophile from terrestrial hot springs in the Azores. International Journal of Systematic and Evolutionary Microbiology , 54, 33-39. Altschul, S., Gish, W., Miller, W., Myers, E., & Lipman, D. (1990). "Basic local alignment search tool.". J. Mol. Biol. , 215:403-410. Amend, J., & Shock, E. (2001). Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and Bacteria. FEMS Microbiology Reviews , 25, 175-243. Aragno, M. (1978). Enrichment, isolation and preliminary characterization of a thermophilic, endospore-forming hydrogen bacterium. FEMS Micobiol. Lett. , 3: 13-15. Aragno, M. (1992). The Thermophilic, Aerobic, Hydrogen-Oxidizing (Knallgas) Bacteria. In A. Balows, H. Trüper, M. Dworkin, W. Harder, & K. Schleifer, The Prokaryotes, a handbook on biology of bacteria. 2nd ed. vol. 4 (pp. 3917-3933.). New York: Springer Verlag. Aragno, M., & Schlegel, H. G. (1992). The mesophilic Hydrogen-Oxidizing (Knallgas) Bacteria. In A. Balows, H. Truper, M. Dworkin, W. Harder, & K.-H. Schleifer, The Prokaryotes 2nd. ed. (pp. 344-384). New York: Springer. Ármannson, H. (2002, May 30-31). Erindi á ráðstefnu um málefni veitufyrirtækja . Grænt bókhald í jarðhita- samanburður á útblæstri við aðra orkugjafa . Akureyri, Iceland: Samorka. Bae, S., Kwak, K., Kim, S., Chung, S., & Igarashi, Y. (2001). Isolation and Characterization of CO2-Fixing Hydrogen -Oxidizing Marine 109 University of Akureyri Department of Natural Resource Science Bacteria. -

Characterization of the Uncommon Enzymes from (2004)
OPEN Mannosylglucosylglycerate biosynthesis SUBJECT AREAS: in the deep-branching phylum WATER MICROBIOLOGY MARINE MICROBIOLOGY Planctomycetes: characterization of the HOMEOSTASIS MULTIENZYME COMPLEXES uncommon enzymes from Rhodopirellula Received baltica 13 March 2013 Sofia Cunha1, Ana Filipa d’Avo´1, Ana Mingote2, Pedro Lamosa3, Milton S. da Costa1,4 & Joana Costa1,4 Accepted 23 July 2013 1Center for Neuroscience and Cell Biology, University of Coimbra, 3004-517 Coimbra, Portugal, 2Instituto de Tecnologia Quı´mica Published e Biolo´gica, Universidade Nova de Lisboa, 2780-157 Oeiras, Portugal, 3Centro de Ressonaˆncia Magne´tica Anto´nio Xavier, 7 August 2013 Instituto de Tecnologia Quı´mica e Biolo´gica, Universidade Nova de Lisboa, 2781-901 Oeiras, Portugal, 4Department of Life Sciences, University of Coimbra, Apartado 3046, 3001-401 Coimbra, Portugal. Correspondence and The biosynthetic pathway for the rare compatible solute mannosylglucosylglycerate (MGG) accumulated by requests for materials Rhodopirellula baltica, a marine member of the phylum Planctomycetes, has been elucidated. Like one of the should be addressed to pathways used in the thermophilic bacterium Petrotoga mobilis, it has genes coding for J.C. ([email protected].) glucosyl-3-phosphoglycerate synthase (GpgS) and mannosylglucosyl-3-phosphoglycerate (MGPG) synthase (MggA). However, unlike Ptg. mobilis, the mesophilic R. baltica uses a novel and very specific MGPG phosphatase (MggB). It also lacks a key enzyme of the alternative pathway in Ptg. mobilis – the mannosylglucosylglycerate synthase (MggS) that catalyses the condensation of glucosylglycerate with GDP-mannose to produce MGG. The R. baltica enzymes GpgS, MggA, and MggB were expressed in E. coli and characterized in terms of kinetic parameters, substrate specificity, temperature and pH dependence. -

Antibiotic Susceptibility of Brachyspira
ACTA VET. BRNO 2014, 83: 003–007; doi:10.2754/avb201483010003 Antibiotic susceptibility of Brachyspira hyodysenteriae isolates from Czech swine farms: a 10-year follow-up study Josef Prášek1, Daniel Šperling2, Dana Lobová1, Jiří Smola2, Alois Čížek1 1University of Veterinary and Pharmaceutical Sciences, Faculty of Veterinary Medicine, Institute of Infectious Diseases and Microbiology, 2Ruminant and Swine Clinic, Brno, Czech Republic Received June 10, 2013 Accepted September 26, 2013 Abstract Brachyspira hyodysenteriae is the causative agent of swine dysentery. Loss of clinical efficacy of some antimicrobial agents authorized for treating swine dysentery was observed on certain Czech pig farms. The aim of the present study was to evaluate the antimicrobial sensitivity of six antibiotics using a set of 202 randomly selected B. hyodysenteriae isolates obtained from farms in the Czech Republic between years 1997 and 2006. Minimum inhibitory concentration of antibiotics tylosin, lincomycin, tylvalosin, chlortetracyclin, tiamulin and valnemulin were tested, using an agar dilution method. All antibiotics tested showed an increase in minimal inhibitory concentrations. Continual decrease in susceptibility of B. hyodysenteriae isolates to tiamulin and valnemulin was observed. Multiresistant B. hyodysenteriae were isolated more frequently in the past years. Only a careful use of antibiotics can ensure their efficacy, especially in case of pleuromutilins, in the strategic therapy of swine dysentery. This rare study demonstrates the minimal inhibitory concentration changes of selected antidysenterics among Czech isolates of Brachyspira hyodysenteriae during a ten-year period. Pigs, swine dysentery, therapy, minimal inhibition concentration, antimicrobial resistance Brachyspira hyodysenteriae is an intestinal spirochete which colonizes the large intestine of pigs after being ingested, and induces diarrhoeal disease – swine dysentery (SD) (Hampson et al. -

Proteome Characterization of Brachyspira Strains
ADVERTIMENT. Lʼaccés als continguts dʼaquesta tesi queda condicionat a lʼacceptació de les condicions dʼús establertes per la següent llicència Creative Commons: http://cat.creativecommons.org/?page_id=184 ADVERTENCIA. El acceso a los contenidos de esta tesis queda condicionado a la aceptación de las condiciones de uso establecidas por la siguiente licencia Creative Commons: http://es.creativecommons.org/blog/licencias/ WARNING. The access to the contents of this doctoral thesis it is limited to the acceptance of the use conditions set by the following Creative Commons license: https://creativecommons.org/licenses/?lang=en Department of Cellular Biology, Physiology and Immunology Doctoral Program in Immunology Proteome characterization of Brachyspira strains. Identification of bacterial antigens. Doctoral Thesis Mª Vanessa Casas López Bellaterra, July 2017 Department of Cellular Biology, Physiology and Immunology Doctoral Program in Immunology Proteome characterization of Brachyspira strains. Identification of bacterial antigens. Doctoral thesis presented by Mª Vanessa Casas López To obtain the Ph.D. in Immunology This work has been carried out in the Proteomics Laboratory CSIC/UAB under the supervision of Dr. Joaquin Abián and Dra. Montserrat Carrascal. Ph.D. Candidate Ph.D. Supervisor Mª Vanessa Casas López Dr. Joaquin Abián Moñux CSIC Research Scientist Department Tutor Ph.D. Supervisor Dra. Dolores Jaraquemada Pérez de Dra. Montserrat Carrascal Pérez Guzmán CSIC Tenured Scientist UAB Immunology Professor Bellaterra, July 2017 “At My Most Beautiful” R.E.M. from the album “Up” (1998) “And after all, you’re my wonderwall” Oasis from the album “(What´s the Story?) Morning Glory” (1995) Agradecimientos A mis directores de tesis, por su tiempo, sus ideas y consejos. -

And Thermo-Adaptation in Hyperthermophilic Archaea: Identification of Compatible Solutes, Accumulation Profiles, and Biosynthetic Routes in Archaeoglobus Spp
Universidade Nova de Lisboa Osmo- andInstituto thermo de Tecnologia-adaptation Química e Biológica in hyperthermophilic Archaea: Subtitle Subtitle Luís Pedro Gafeira Gonçalves Osmo- and thermo-adaptation in hyperthermophilic Archaea: identification of compatible solutes, accumulation profiles, and biosynthetic routes in Archaeoglobus spp. OH OH OH CDP c c c - CMP O O - PPi O3P P CTP O O O OH OH OH OH OH OH O- C C C O P O O P i Dissertation presented to obtain the Ph.D degree in BiochemistryO O- Instituto de Tecnologia Química e Biológica | Universidade Nova de LisboaP OH O O OH OH OH Oeiras, Luís Pedro Gafeira Gonçalves January, 2008 2008 Universidade Nova de Lisboa Instituto de Tecnologia Química e Biológica Osmo- and thermo-adaptation in hyperthermophilic Archaea: identification of compatible solutes, accumulation profiles, and biosynthetic routes in Archaeoglobus spp. This dissertation was presented to obtain a Ph. D. degree in Biochemistry at the Instituto de Tecnologia Química e Biológica, Universidade Nova de Lisboa. By Luís Pedro Gafeira Gonçalves Supervised by Prof. Dr. Helena Santos Oeiras, January, 2008 Apoio financeiro da Fundação para a Ciência e Tecnologia (POCI 2010 – Formação Avançada para a Ciência – Medida IV.3) e FSE no âmbito do Quadro Comunitário de apoio, Bolsa de Doutoramento com a referência SFRH / BD / 5076 / 2001. ii ACKNOWNLEDGMENTS The work presented in this thesis, would not have been possible without the help, in terms of time and knowledge, of many people, to whom I am extremely grateful. Firstly and mostly, I need to thank my supervisor, Prof. Helena Santos, for her way of thinking science, her knowledge, her rigorous criticism, and her commitment to science. -

Brachyspira (Serpulina) Pilosicoli and Intestinal Spirochetosis: How DIAGNOSTIC NOTES Much Do We Know? Swine Health Prod
Stevenson GW. Brachyspira (Serpulina) pilosicoli and intestinal spirochetosis: How DIAGNOSTIC NOTES much do we know? Swine Health Prod. 1999;7(6):287–291. Brachyspira (Serpulina) pilosicoli and intestinal spirochetosis: How much do we know? Gregory W. Stevenson, DVM, PhD, Diplomate ACVP here are at least five distinct species of Brachyspira nus name based on historic precedent. The genus Brachyspira was es- (Serpulina) known to infect the large intestine of swine.1–3 tablished when Brachyspira aalborgi was first described,7 which oc- Two species are pathogenic: curred prior to the establishment of the genus Serpulina.5 Unfortu- nately, Serpulina intermedia and murdochii were not included in the • Brachyspira hyodysenteriae (formerly Serpulina or Treponema comparative study. For now, they remain in the genus Serpulina. hyodysenteriae), which causes swine dysentery; and • Brachyspira pilosicoli (formerly Serpulina pilosicoli or Brachyspira pilosicoli Anguillina coli), which causes intestinal spirochetosis. Brachyspira pilosicoli can be presumptively differentiated from other Three additional species are nonpathogenic: Brachyspira (Serpulina) spp. by culture (weak β-hemolysis) and biochemical testing. Brachyspira pilosicoli is indole negative and hip- • Brachyspira innocens, formerly Serpulina or Treponema purate-hydrolysis positive, and lack β-glucosidase activity in the API- innocens ZYM profile.11,12 Definitive identification of B. pilosicoli requires PCR • Serpulina intermedia testing.12,13–15 The medium that is most commonly used to culture B. • Serpulina murdochii hyodysenteriae in diagnostic laboratories, BJ medium,16 is slightly in- Selected characteristics of each species are summarized in Table 1. hibitory when used to isolate B. pilosicoli, due to the moderate sensi- tivity of B. pilosicoli to two of the included antibiotics, rifampicin, and At a light microscopic level, all five of these organisms are morphologi- spiramycin.17 Culture of B. -

Methods for Improving Diagnostic Techniques Used for the Identification and Isolation of Brachyspira Species from Swine Hallie Warneke Iowa State University
Iowa State University Capstones, Theses and Graduate Theses and Dissertations Dissertations 2017 Methods for improving diagnostic techniques used for the identification and isolation of Brachyspira species from swine Hallie Warneke Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/etd Part of the Animal Diseases Commons Recommended Citation Warneke, Hallie, "Methods for improving diagnostic techniques used for the identification and isolation of Brachyspira species from swine" (2017). Graduate Theses and Dissertations. 15453. https://lib.dr.iastate.edu/etd/15453 This Thesis is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. Methods for improving diagnostic techniques used for the identification and isolation of Brachyspira species from swine by Hallie L Warneke A thesis submitted to the graduate faculty in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE Major: Veterinary Preventive Medicine Program of Study Committee: Eric R Burrough, Major Professor Timothy S Frana Annette M O’Connor The student author and the program of study committee are solely responsible for the content of this thesis. The Graduate College will ensure this thesis is globally accessible and will not permit alterations after a degree is conferred. Iowa State University Ames, Iowa 2017 Copyright © Hallie L Warneke, 2017. All rights reserved. ii TABLE OF CONTENTS Page LIST OF FIGURES ................................................................................................... iii LIST OF TABLES ....................................................................................................