Haloalkane Dehalogenases from Marine Organisms
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Chapter 7, Haloalkanes, Properties and Substitution Reactions of Haloalkanes Table of Contents 1
Chapter 7, Haloalkanes, Properties and Substitution Reactions of Haloalkanes Table of Contents 1. Alkyl Halides (Haloalkane) 2. Nucleophilic Substitution Reactions (SNX, X=1 or 2) 3. Nucleophiles (Acid-Base Chemistry, pka) 4. Leaving Groups (Acid-Base Chemistry, pka) 5. Kinetics of a Nucleophilic Substitution Reaction: An SN2 Reaction 6. A Mechanism for the SN2 Reaction 7. The Stereochemistry of SN2 Reactions 8. A Mechanism for the SN1 Reaction 9. Carbocations, SN1, E1 10. Stereochemistry of SN1 Reactions 11. Factors Affecting the Rates of SN1 and SN2 Reactions 12. --Eliminations, E1 and E2 13. E2 and E1 mechanisms and product predictions In this chapter we will consider: What groups can be replaced (i.e., substituted) or eliminated The various mechanisms by which such processes occur The conditions that can promote such reactions Alkyl Halides (Haloalkane) An alkyl halide has a halogen atom bonded to an sp3-hybridized (tetrahedral) carbon atom The carbon–chlorine and carbon– bromine bonds are polarized because the halogen is more electronegative than carbon The carbon-iodine bond do not have a per- manent dipole, the bond is easily polarizable Iodine is a good leaving group due to its polarizability, i.e. its ability to stabilize a charge due to its large atomic size Generally, a carbon-halogen bond is polar with a partial positive () charge on the carbon and partial negative () charge on the halogen C X X = Cl, Br, I Different Types of Organic Halides Alkyl halides (haloalkanes) sp3-hybridized Attached to Attached to Attached -

Conformational Landscapes of Halohydrin Dehalogenases and Their Accessible Active Site Tunnels
catalysts Article Conformational Landscapes of Halohydrin Dehalogenases and Their Accessible Active Site Tunnels 1 1, , 1,2, Miquel Estévez-Gay , Javier Iglesias-Fernández * y and Sílvia Osuna * 1 CompBioLab Group, Institut de Química Computacional i Catàlisi (IQCC) and Departament de Química, Universitat de Girona, c/Maria Aurèlia Capmany 69, 17003 Girona, Catalonia, Spain; [email protected] 2 ICREA, Passeig Lluís Companys 23, 08010 Barcelona, Catalonia, Spain * Correspondence: [email protected] (J.I.-F.); [email protected] (S.O.) Present address: Nostrum Biodiscovery, Carrer de Baldiri Reixac, 10–12, 08028 Barcelona, Catalonia, Spain. y Received: 12 November 2020; Accepted: 28 November 2020; Published: 1 December 2020 Abstract: Halohydrin dehalogenases (HHDH) are industrially relevant biocatalysts exhibiting a promiscuous epoxide-ring opening reactivity in the presence of small nucleophiles, thus giving access to novel carbon–carbon, carbon–oxygen, carbon–nitrogen, and carbon–sulfur bonds. Recently, the repertoire of HHDH has been expanded, providing access to some novel HHDH subclasses exhibiting a broader epoxide substrate scope. In this work, we develop a computational approach based on the application of linear and non-linear dimensionality reduction techniques to long time-scale Molecular Dynamics (MD) simulations to study the HHDH conformational landscapes. We couple the analysis of the conformational landscapes to CAVER calculations to assess their impact on the active site tunnels and potential ability towards bulky epoxide ring opening reaction. Our study indicates that the analyzed HHDHs subclasses share a common breathing motion of the halide binding pocket, but present large deviations in the loops adjacent to the active site pocket and N-terminal regions. -

Molecular Basis of Two Novel Dehalogenating Activities in Bacteria
Molecular basis of two novel dehalogenating activities in bacteria Structure/function relationships in an evolutionary context 1 Foto’s omslag: Dinanda Lutikhedde, MFA Druk: Digital Printing Partners Utrecht bv The research presented in this work was performed in the protein crystallography group at the Laboratory of Biophysical Chemistry of the University of Groningen. This work was supported by The Netherlands Foundation of Chemical Research (CW) with financial aid from The Netherlands Foundation for Scientific Research (NWO). 2 RIJKSUNIVERSITEIT GRONINGEN Molecular basis of two novel dehalogenating activities in bacteria Structure/function relationships in an evolutionary context Proefschrift ter verkrijging van het doctoraat in de Wiskunde en Natuurwetenschappen aan de Rijksuniversiteit Groningen op gezag van de Rector Magnificus, dr. F. Zwarts, in het openbaar te verdedigen op vrijdag 30 januari 2004 om 14.15 uur door René Marcel de Jong geboren op 22 maart 1976 te Heerenveen 3 Promotor: Prof. dr. B. W. Dijkstra Beoordelingscommissie: Prof. dr. D. B. Janssen Prof. dr. A. E. Mark Prof. dr. J.B.F.N. Engberts 4 CONTENTS CHAPTER 1: GENERAL INTRODUCTION ..........................................................9 EVOLUTION IN THE MICROBIAL WORLD.................................................................................. 10 EVOLUTION AT THE MOLECULAR LEVEL................................................................................. 12 BACTERIAL DEHALOGENATING ACTIVITY TOWARDS ANTHROPOGENIC COMPOUNDS ................... 14 SCOPE -

Classifying Halogenoalkanes Reactions Of
10 Halogenoalkanes H Naming Halogenoalkanes H H H H C H H H H Based on original alkane, with a prefix indicating halogen atom: H C C C H Fluoro for F; Chloro for Cl; Bromo for Br; Iodo for I. H C C C C H Br H H H Cl H H Substituents are listed alphabetically 1-bromopropane 2-chloro-2-methylbutane Classifying Halogenoalkanes Halogenoalkanes can be classified as primary, secondary or tertiary depending on the number of carbon atoms attached to the C-X functional group. H H H H H H H H C H H H H H C C C H H C C C H H C C C C H Br H H H Br H H Cl H H Primary halogenoalkane Secondary halogenoalkane Tertiary halogenoalkane One carbon attached to the Two carbons attached to the Three carbons attached to the carbon atom adjoining the carbon atom adjoining the carbon atom adjoining the halogen halogen halogen Reactions of Halogenoalkanes Halogenoalkanes undergo either substitution or elimination reactions Nucleophilic substitution reactions Substitution: swapping a halogen atom for another atom or groups of atoms - - Nucleophile: electron pair donator e.g. :OH , :NH3, CN :Nu represents any nucleophile – they The Mechanism: We draw (or outline) mechanisms to always have a lone pair and act as show in detail how a reaction proceeds electron pair donators Nu:- The nucleophiles The carbon has a small attack the positive H H H H positive charge because carbon atom of the electronegativity H C C X H C C + X- δ+ δ- Nu difference between the carbon and the halogen H H H H We use curly arrows in mechanisms (with A curly arrow will always two line heads) to show the movement of start from a of electrons or two electrons lone pair the centre of a bond The rate of these substitution reactions depends on the strength Bond enthalpy / of the C-X bond kJmol-1 The weaker the bond, the easier it is to break and the faster the reaction. -
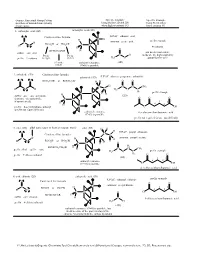
Generic Functional Group Pattern in Ordero of Nomenclature Priority in Our Course. Specific Example Using Suffix (2D and 3D)
Generic Functional Group Pattern Specific Example Specific Example in ordero of nomenclature priority Using Suffix (2D and 3D) Using Prefix when in our course. when highest priority FG lower priority FG 1. carboxylic acid (2D) carboxylic acid (3D) O Condensed line formula IUPAC: ethanoic acid O H H prefix example C H common: acetic acid RCO2H or HO2CR C C R O #-carboxy H O RCH(CO H)R' H suffix: -oic acid 2 O not used in our course FG on FG on C H (acids are the highest priority prefix: #-carboxy the right the left H3C O group that we use) FG in the carbonyl resonance (2D) middle (C=O) is possible 2. anhydride (2D) Condensed line formula anhydride (3D) IUPAC: ethanoic propanoic anhydride O O RCO2COR' or ROCO2CR' H H O O H C C C CH C 3 R O R H3C O C H H 2 prefix example H O C H suffix: -oic -oic anhydride (2D) (just one -oic anhydride, O O O C C O if symmetrical) 3 1 H 4 H 2 prefix: #-acyloxyalkanecarbonyl O O OH (prefix not required for us) carbonyl resonance 4-acyloxymethanebutanoic acid (C=O) is possible (prefix not required for us - too difficult) 3. ester (2D) alkyl name (goes in front as separate word) ester (3D) H H H IUPAC: propyl ethanoate O C Condensed line formula C O C common: propyl acetate C R' H H R O RCO2R' or R'O2CR H H C C H O H H2 RCH(CO2CH3)R' prefix: alkyl suffix: -oate H C C CH3 O H C O C prefix example 3 H prefix: #-alkoxycarbonyl 2 O O (2D) 3 1 carbonyl resonance 4 2 (C=O) is possible O OH 4-methoxycarbonylbutanoic acid 4. -

A Perovskite-Based Paper Microfluidic Sensor for Haloalkane Assays
ORIGINAL RESEARCH published: 26 April 2021 doi: 10.3389/fchem.2021.682006 A Perovskite-Based Paper Microfluidic Sensor for Haloalkane Assays Lili Xie 1, Jie Zan 1, Zhijian Yang 1, Qinxia Wu 1, Xiaofeng Chen 1, Xiangyu Ou 1, Caihou Lin 2*, Qiushui Chen 1,3* and Huanghao Yang 1,3* 1 Ministry of Education (MOE) Key Laboratory for Analytical Science of Food Safety and Biology, Fujian Provincial Key Laboratory of Analysis and Detection Technology for Food Safety, College of Chemistry, Fuzhou University, Fuzhou, China, 2 Department of Neurosurgery, Fujian Medical University Union Hospital, Fuzhou, China, 3 Fujian Science and Technology Innovation Laboratory for Optoelectronic Information of China, Fuzhou, China Detection of haloalkanes is of great industrial and scientific importance because some Edited by: haloalkanes are found serious biological and atmospheric issues. The development Jinyi Lin, of a flexible, wearable sensing device for haloalkane assays is highly desired. Here, Nanjing Tech University, China we develop a paper-based microfluidic sensor to achieve low-cost, high-throughput, Reviewed by: Xiaobao Xu, and convenient detection of haloalkanes using perovskite nanocrystals as a nanoprobe Nanjing University of Science and through anion exchanging. We demonstrate that the CsPbX3 (X = Cl, Br, or I) Technology, China nanocrystals are selectively and sensitively in response to haloalkanes (CH2Cl2, CH2Br2), Xuewen Wang, Northwestern Polytechnical and their concentrations can be determined as a function of photoluminescence University, China spectral shifts of perovskite nanocrystals. In particular, an addition of nucleophilic trialkyl *Correspondence: phosphines (TOP) or a UV-photon-induced electron transfer from CsPbX3 nanocrystals is Caihou Lin [email protected] responsible for achieving fast sensing of haloalkanes. -

Reactions of Alkenes and Alkynes
05 Reactions of Alkenes and Alkynes Polyethylene is the most widely used plastic, making up items such as packing foam, plastic bottles, and plastic utensils (top: © Jon Larson/iStockphoto; middle: GNL Media/Digital Vision/Getty Images, Inc.; bottom: © Lakhesis/iStockphoto). Inset: A model of ethylene. KEY QUESTIONS 5.1 What Are the Characteristic Reactions of Alkenes? 5.8 How Can Alkynes Be Reduced to Alkenes and 5.2 What Is a Reaction Mechanism? Alkanes? 5.3 What Are the Mechanisms of Electrophilic Additions HOW TO to Alkenes? 5.1 How to Draw Mechanisms 5.4 What Are Carbocation Rearrangements? 5.5 What Is Hydroboration–Oxidation of an Alkene? CHEMICAL CONNECTIONS 5.6 How Can an Alkene Be Reduced to an Alkane? 5A Catalytic Cracking and the Importance of Alkenes 5.7 How Can an Acetylide Anion Be Used to Create a New Carbon–Carbon Bond? IN THIS CHAPTER, we begin our systematic study of organic reactions and their mecha- nisms. Reaction mechanisms are step-by-step descriptions of how reactions proceed and are one of the most important unifying concepts in organic chemistry. We use the reactions of alkenes as the vehicle to introduce this concept. 129 130 CHAPTER 5 Reactions of Alkenes and Alkynes 5.1 What Are the Characteristic Reactions of Alkenes? The most characteristic reaction of alkenes is addition to the carbon–carbon double bond in such a way that the pi bond is broken and, in its place, sigma bonds are formed to two new atoms or groups of atoms. Several examples of reactions at the carbon–carbon double bond are shown in Table 5.1, along with the descriptive name(s) associated with each. -
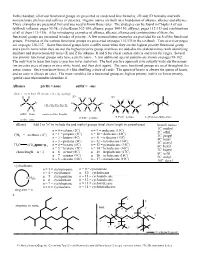
In This Handout, All of Our Functional Groups Are Presented As Condensed Line Formulas, 2D and 3D Formulas and with Nomenclature Prefixes and Suffixes (If Present)
In this handout, all of our functional groups are presented as condensed line formulas, 2D and 3D formulas and with nomenclature prefixes and suffixes (if present). Organic names are built on a foundation of alkanes, alkenes and alkynes. Those examples are presented first and you need to know those rules. The strategies can be found in Chapter 4 of our textbook (alkanes: pages 93-98, cycloalkanes 102-104, alkenes: pages 104-110, alkynes: pages 112-113 and combinations of all of them 113-115). After introducing examples of alkanes, alkenes, alkynes and combinations of them, the functional groups are presented in order of priority. A few nomenclature examples are provided for each of the functional groups. Examples of the various functional groups are presented on pages 115-135 in the textbook. Two overview pages are on pages 136-137. Some functional groups have a suffix name when they are the highest priority functional group and a prefix name when they are not the highest priority group, and these are added to the skeletal names with identifying numbers and stereochemistry terms (E and Z for alkenes, R and S for chiral centers and cis and trans for rings). Several low priority functional groups only have a prefix name. A few additional special patterns are shown on pages 98-102. The only way to learn this topic is practice (over and over). The best practice approach is to actually write out the names (on an extra piece of paper or on a white board, and then do it again). The same functional groups are used throughout the entire course. -

Fully Halogenated Chlorofluorocarbons
INTERNATIONAL PROGRAMME ON CHEMICAL SAFETY ENVIRONMENTAL HEALTH CRITERIA 113 FULLY HALOGENATED CHLOROFLUOROCARBONS This report contains the collective views of an international group of experts and does not necessarily represent the decisions or the stated policy of the United Nations Environment Programme, the International Labour Organisation, or the World Health Organization. Published under the joint sponsorship of the United Nations Environment Programme, the International Labour Organisation, and the World Health Organization World Health Orgnization Geneva, 1990 The International Programme on Chemical Safety (IPCS) is a joint venture of the United Nations Environment Programme, the International Labour Organisation, and the World Health Organization. The main objective of the IPCS is to carry out and disseminate evaluations of the effects of chemicals on human health and the quality of the environment. Supporting activities include the development of epidemiological, experimental laboratory, and risk-assessment methods that could produce internationally comparable results, and the development of manpower in the field of toxicology. Other activities carried out by the IPCS include the development of know-how for coping with chemical accidents, coordination of laboratory testing and epidemiological studies, and promotion of research on the mechanisms of the biological action of chemicals. WHO Library Cataloguing in Publication Data Fully halogenated chlorofluorocarbons. (Environmental health criteria ; 113) 1.Freons - adverse effects 2.Freons - toxicity I.Series ISBN 92 4 157113 6 0 (NLM Classification: QV 633) ISSN 0250-863X The World Health Organization welcomes requests for permission to reproduce or translate its publications, in part or in full. Applications and enquiries should be addressed to the Office of Publications, World Health Organization, Geneva, Switzerland, which will be glad to provide the latest information on any changes made to the text, plans for new editions, and reprints and translations already available. -

(12) United States Patent (10) Patent No.: US 7,381,538 B2 Reardon Et Al
US007381538B2 (12) United States Patent (10) Patent No.: US 7,381,538 B2 Reardon et al. (45) Date of Patent: Jun. 3, 2008 (54) OPTICAL BIOSENSOR WITH ENHANCED 6,060,327 A * 5/2000 Keen ..................... 204,403.14 ACTIVITY RETENTION FOR DETECTION 6,159,681 A 12/2000 Zebala .......................... 435/4 OF HALOGENATED ORGANIC COMPOUNDS OTHER PUBLICATIONS Derek W. Campbell, entitled “The Development of Biosensors for (75) Inventors: Kenneth F. Reardon, Fort Collins, CO the Detection of Halogenated Groundwater Contaminants,” avail (US); Neema Das, Murgeshpalya (IN) able from Morgan Library at the CSU in Fort Collins, Colorado, having been submitted by Derek W. Campbell in fulfillment of the (73) Assignee: Colorado State University Research requirements for the Degree of Master of Science at Colorado State Foundation (CSURF), Fort Collins, University, Spring 1998 (labeled “Attachment B” of applicants' CO (US) provisional app. filed Jun. 1, 2001 as containing general background technical information}. (*) Notice: Subject to any disclaimer, the term of this inst k Eyle A. PS SEE s past l S.s I listed under 35 12,alogenated 1998 by Reardonflydrocarbons, and Campbell, U.S. Appl. 7 pages No. (labeled,89U, “Attachment Illed Sep. M YW- (b) by ayS. C” of applicants’pp pprovisional app.pp filed Jun. 1, 2001 as containin9. general background technical information}. (21) Appl. No.: 10/478,822 Cord Müller. F. Schubert. T. Scheper,p "Multicomponentp fiberopticalp biosensor for use in hemodialysis monitoring.” Proc. SPIE vol. (22) PCT Filed: Jun. 1, 2002 2131, 555-562 Biomedical Fiber Optic Instrumentation(Jul 1994) {referenced as “Cord Müller, et al.” in applicants’ provisional app. -

Haloalkanes, Alcohols and Amines. Year 1 Dr
Haloalkanes, Alcohols and Amines. Year 1 Dr. Chris Braddock [[email protected], Rm 440]: 10 lectures Aims of course: To introduce the functional group chemistry of haloalkanes, alcohols and amines, building on the functional group approach of Dr Smith's course where alkanes, alkenes, alkynes and benzene were discussed. Course objectives: At the end of this course you should be able to: • Identify correctly the various functional groups and reagents introduced on this course; • Recognise characteristic IR and NMR signals in their spectra; • Predict the product of a reaction involving an haloalkane, alcohol or amine with a given reagent; • Select reagents for the common transformations of haloalkanes, alcohols and amines into other functionalities; • Select reagents for the preparation of haloalkanes, alcohols and amines from other substrates; • Explain the mechanistic rationale underpinning the above. Recommended Texts Vollhardt and Schore, Organic Chemistry, 3rd edition, WH Freeman & Co, New York, 1999. Sykes A Primer to Mechanism in Organic Chemistry Longman, Harlow, 1995 Clayden, Greeves, Warren and Wothers Organic Chemistry OUP, Oxford 2001. Course Content A. Haloalkane Group Chemistry (Alkyl Halides) (a) Structure and Physical Properties: nomenclature; physical properties; spectroscopic properties. β (b) Haloalkane group reactivity: Nucleophilic substitution S N1 and SN2; - elimination; metal-halogen exchange. (c) Synthesis of haloalkanes: electrophilic addition to alkenes; radical addition to alkenes; from alcohols. B. Hydroxyl Group Chemistry (Alcohols) (a) Structure and Physical Properties: Structure; H-bonding; spectroscopic properties . (b) Alcohol group reactivity: Hydroxyl group acidity; Nucleophilic reactions of hydroxyl groups without C-O cleavage; nucleophilic reactions of hydroxyl groups which convert the OH into a good leaving group; nucleophilic reactions of hydroxyl groups which place a good leaving group on oxygen. -

Haloalkanes and Haloarenes
1010Unit Objectives HaloalkanesHaloalkanes andand After studying this Unit, you will be able to HaloarHaloarHaloarHaloarenesenesenesenesenesenes • name haloalkanes and haloarenes according to the IUPAC system of nomenclature from their given structures; Halogenated compounds persist in the environment due to their • describe the reactions involved in resistance to breakdown by soil bacteria. the preparation of haloalkanes and haloarenes and understand various reactions that they The replacement of hydrogen atom(s) in an aliphatic undergo; or aromatic hydrocarbon by halogen atom(s) results • correlate the structures of in the formation of alkyl halide (haloalkane) and aryl haloalkanes and haloarenes with halide (haloarene), respectively. Haloalkanes contain various types of reactions; halogen atom(s) attached to the sp3 hybridised carbon • use stereochemistry as a tool for atom of an alkyl group whereas haloarenes contain understanding the reaction halogen atom(s) attached to sp2 hybridised carbon mechanism; atom(s) of an aryl group. Many halogen containing • appreciate the applications of organic compounds occur in nature and some of organo-metallic compounds; these are clinically useful. These classes of compounds • highlight the environmental effects of polyhalogen compounds. find wide applications in industry as well as in day- to-day life. They are used as solvents for relatively non-polar compounds and as starting materials for the synthesis of wide range of organic compounds. Chlorine containing antibiotic, chloramphenicol, produced by microorganisms is very effective for the treatment of typhoid fever. Our body produces iodine containing hormone, thyroxine, the deficiency of which causes a disease called goiter. Synthetic halogen compounds, viz. chloroquine is used for the treatment of malaria; halothane is used as an anaesthetic during surgery.