Variations of Glossina Sp. and Trypanosome Species Frequency Within Different Habitats in a Sleeping Sickness Focus, Gabon
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
C:\Hnp2\Gabon00\ASSETS.DAT
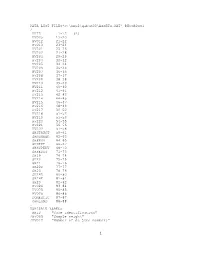
DATA LIST FILE='c:\hnp2\gabon00\ASSETS.DAT' RECORDS=1 / HHID 1-12 (A) HV005 13-20 HV012 21-22 HV013 23-24 HV201 25-26 HV202 27-28 HV203 29-29 HV204 30-32 HV205 33-34 HV206 35-35 HV207 36-36 HV208 37-37 HV209 38-38 HV210 39-39 HV211 40-40 HV212 41-41 HV213 42-43 HV214 44-45 HV215 46-47 HV216 48-49 HV217 50-50 HV218 51-52 HV219 53-53 HV220 54-55 HV221 56-56 HV222 57-58 SHSTRUCT 59-61 SHNUMBER 62-63 SHPROV 64-65 SHDEPT 66-67 SHSUPERV 68-70 SHFEDIT 71-73 SH19 74-74 SH20 75-75 SH21 76-76 SH22D 77-77 SH23 78-79 SH24D 80-80 SH24E 81-81 SH29 82-82 HV024 83-84 HV025 85-85 HV026 86-86 DOMESTIC 87-87 OWNLAND 88-88 . VARIABLE LABELS HHID "Case Identification" /HV005 "Sample weight" /HV012 "Number of de jure members" 1 /HV013 "Number of de facto members" /HV201 "Source of drinking water" /HV202 "Source of non-drinking wate-NA" /HV203 "Same source of water -NA" /HV204 "Time to get to water source" /HV205 "Type of toilet facility" /HV206 "Has electricity" /HV207 "Has radio" /HV208 "Has television" /HV209 "Has refrigerator" /HV210 "Has bicycle" /HV211 "Has motorcycle" /HV212 "Has car" /HV213 "Main floor material" /HV214 "Main wall material" /HV215 "Main roof material" /HV216 "Rooms for sleeping" /HV217 "Relationship structure" /HV218 "Line number of head of househ." /HV219 "Sex of head of household" /HV220 "Age of head of household" /HV221 "Has telephone" /HV222 "Type of salt used for cooking" /SHSTRUCT "Structure number" /SHNUMBER "Household number within a structure" /SHPROV "Province" /SHDEPT "Departement" /SHSUPERV "Supervisor code" /SHFEDIT "Field editor code" /SH19 "Distance between house and toilets" /SH20 "Latrine depth" /SH21 "Toilets shared with others" /SH22D "Has video" /SH23 "Fuel used in the household" /SH24D "Boat without engine" /SH24E "Powered boat" /SH29 "Salt test result" /HV024 "Region" /HV025 "Type of place of residence" /HV026 "Place of residence" /DOMESTIC "If HH has a domestic worker not related to head" /OWNLAND "If household works own or family's agric. -

Plan De Développement Stratégique De La Fenatag - 2018-2022
RÉPUBLIQUE DU GABON FÉDÉRATION NATIONALE DES TRANSFORMATEURS DES PRODUITS AGRICOLES DU GABON PLAN DE DÉVELOPPEMENT STRATÉGIQUE DE LA FENATAG - 2018-2022 RÉPUBLIQUE DU GABON FÉDÉRATION NATIONALE DES TRANSFORMATEURS DES PRODUITS AGRICOLES DU GABON PLAN DE DÉVELOPPEMENT STRATÉGIQUE DE LA FENATAG - 2018-2022 Organisation des Nations Unies pour l’alimentation et l’agriculture Libreville, 2018 3 Les appellations employées dans ce produit d’information et la présentation des données qui y figurent n’impliquent de la part de l’Organisation des Nations Unies pour l’alimentation et l’agriculture (FAO) aucune prise de position quant au statut juridique ou au stade de développement des pays, territoires, villes ou zones ou de leurs autorités, ni quant au tracé de leurs frontières ou limites. La mention de sociétés déterminées ou de produits de fabricants, qu’ils soient ou non brevetés, n’entraîne, de la part de la FAO, aucune approbation ou recommandation desdits produits de préférence à d’autres de nature analogue qui ne sont pas cités. Les opinions exprimées dans ce produit d’information sont celles du/des auteur(s) et ne reflètent pas nécessairement les vues ou les politiques de la FAO. ISBN 978-92-5-130772-4 © FAO, 2018 La FAO encourage l’utilisation, la reproduction et la diffusion des informations figurant dans ce produit d’information. Sauf indication contraire, le contenu peut être copié, téléchargé et imprimé aux fins d’étude privée, de recherches ou d’enseignement, ainsi que pour utilisation dans des produits ou services non commerciaux, sous réserve que la FAO soit correctement mentionnée comme source et comme titulaire du droit d’auteur et à condition qu’il ne soit sous-en - tendu en aucune manière que la FAO approuverait les opinions, produits ou services des utilisateurs. -

Strategie Nationale D'adaptation Du Littoral
Stratégie Nationale d’Adaptation du littoral gabonais face aux effets des changements climatiques STRATEGIE NATIONALE D’ADAPTATION DU LITTORAL GABONAIS FACE AUX EFFETS DES CHANGEMENTS CLIMATIQUES DOCUMENT Nº 1 INTRODUCTION ET METHODOLOGIE FONDS POUR L’ENVIRONNEMENT MONDIAL Du PROGRAMME DES NATIONS UNIES POUR LE DEVELOPPEMENT (PNUD) Projet A.A.P.: Renforcement des capacités institutionnelles pour une meilleure adaptation en zone côtière au Gabon. Octobre 2011 Document nº 1. Introduction et méthodologie Stratégie Nationale d’Adaptation du littoral gabonais face aux effets des changements climatiques SOMMAIRE 1. ANTÉCÉDENTS .................................................................................... 1 2. MOTIF .................................................................................................... 3 3. INTRODUCTION .................................................................................. 4 3.1. INITIATIVES INTERNATIONALES DANS LA GESTION DE L’ESPACE LITTORAL ................................................................................. 7 3.2. INITIATIVES AU GABON .......................................................................... 22 4. METHODOLOGIE ............................................................................. 29 4.1. OBJECTIFS ................................................................................................... 32 4.1.1. De la Stratégie .............................................................................................. 32 4.1.2. Du travail ..................................................................................................... -

Report of a WHO Meeting on Elimination of African Trypanosomiasis (Trypanosoma Brucei Gambiense)
Report of a WHO meeting on elimination of African trypanosomiasis (Trypanosoma brucei gambiense) Geneva, 3–5 December 2012 « Le but n’est pas toujours placé pour être atteint mais pour servir de point de mire » “The goal is not always meant to be reached, but to serve as a mark for our aim.” Joseph Joubert, 1754–1824 © World Health Organization 2013 All rights reserved. Publications of the World Health Organization are available on the WHO web site (www.who.int) or can be purchased from WHO Press, World Health Organization, 20 Avenue Appia, 1211 Geneva 27, Switzerland (tel.: +41 22 791 3264; fax: +41 22 791 4857;e-mail: [email protected]) Requests for permission to reproduce or translate WHO publications –whether for sale or for non-commercial distribution– should be addressed to WHO Press through the WHO web site (www.who.int/about/licensing/copyright_form/en/index.html). The designations employed and the presentation of the material in this publication do not imply the expression of any opinion whatsoever on the part of the World Health Organization concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. Dotted lines on maps represent approximate border lines for which there may not yet be full agreement. The mention of specific companies or of certain manufacturers’ products does not imply that they are endorsed or recommended by the World Health Organization in preference to others of a similar nature that are not mentioned. Errors and omissions excepted, the names of proprietary products are distinguished by initial capital letters. -

Instruction Comptable Janvier20
MINISTERE DU BUDGET REPUBLIQUE GABONAISE ET DES COMPTES PUBLICS --------------- --------------- Union - Travail - Justice SECRETARIAT GENERAL -------------- DIRECTION GENERALE DE LA COMPTABILITE PUBLIQUE ET DU TRESOR ------------- INSTRUCTION SUR LA COMPTABILITE DE L’ETAT Janvier 2017 MINISTERE DU BUDGET REPUBLIQUE GABONAISE ET DES COMPTES PUBLICS --------------- --------------- Union - Travail - Justice SECRETARIAT GENERAL -------------- DIRECTION GENERALE DE LA COMPTABILITE PUBLIQUE ET DU TRESOR ------------- INSTRUCTION SUR LA COMPTABILITE DE L’ETAT Janvier 2017 Comptabilité de l’Etat 2 Instruction sur la ccomptabilité de l’Etat Comptabilité de l’Etat 4 SOMMAIRE LIVRE 1 : DISPOSITIONS GENERALES ............................................................................................................................ 7 TITRE 1 : L’ORGANISATION DE LA DIRECTION GENERALE DE LA COMPTABILITE PUBLIQUE ET DU TRESOR...................... 9 TITRE 2 : L’ORGANISATION DE LA FONCTION COMPTABILITE ET LES PERIODES COMPTABLES ...................................... 23 LIVRE 2 : COMPTABILISATION DES OPERATIONS DE L’ETAT ....................................................................................... 35 TITRE 1: LA COMPTABILISATION DES OPERATIONS DE RECETTES ................................................................................... 37 TITRE 2: COMPTABILISATION DES OPERATIONS DE DEPENSE ET DE FINANCEMENT ...................................................... 65 TITRE 3: LES OPERATIONS DE TRESORERIE .................................................................................................................... -

Republique Gabonaise Union- Travail – Justice
REPUBLIQUE GABONAISE UNION- TRAVAIL – JUSTICE MINISTERE DE LA SANTE PLAN DIRECTEUR DE LUTTE CONTRE LES MALADIES TROPICALES NEGLIGEES Avec l’appui de novembre 2012 i ABREVIATIONS ET ACRONYMES Abréviations Signification AAEP : Assainissement et Approvisionnement en eau potable ACD : Approche pour atteindre chaque département AFRO : Bureau Régional pour l’Afrique de l’Organisation mondiale de la Santé ALB : Albendazole AMS : Assemblée Mondiale de la Santé APOC : Programme Africain de Lutte contre l’Onchocercose BAAR : Bacille Acido Alcoolo Résistant BCG : Bacille de Calmette et Guérin BELE : Base d'Epidémiologie et de Lutte contre les Endémies CDT : Centre de Diagnostic et de Traitement CEEAC Communauté des Etats de l’Afrique Centrale CEMAC : Communauté Economique et Monétaire de l’Afrique Centrale CHDs : Journées de la santé de l’enfant (= Children health Days) CHU / CHR : Centre Hospitalier Universitaire / Centre Hospitalier Régional CHANGES : Appui en santé communautaire et nutrition, équité sexuelle et éducation CIRMF : Centre International de Recherches Médicales des Franceville CICIBA : Centre International de Civilisation Bantoue CM / CS : Centre Médical / Centre de Santé CNAMGS : Caisse Nationale d’Assurance Maladie et de Garantie Sociale CNSS : Caisse Nationale de Sécurité Sociale COSP : Cellule d'Observation en Santé Publique CTP : Chimiothérapie préventive DIS : Direction de l'Informatique et des Statistiques DALY : Années de vie ajustées sur l’incapacité DMP : Direction du Médicament et de la Pharmacie DMM : Distribution de Masse -
Urban Transmission of Human African Trypanosomiasis, Gabon
LETTERS at symptom onset, but results for Acknowledgments 8. Valiente Moro C, De Luna CJ, Tod A, patient serum samples cultured We thank V. Rupeš for parasitologic Guy JH, Sparagano OAE, Zenner L. The poultry red mite (Dermanyssus gallinae): under the same conditions as the analysis, A. Valkoun for serologic analysis a potential vector of pathogenic agents. homogenized parasites remained of specifi c antibodies to Rickettsia and Exp Appl Acarol. 2009;48:93–104. negative. Signifi cant titers of IgG Coxiella spp., D. Kafková for collection doi:10.1007/s10493-009-9248-0 against B. quintana and B. henselae of patient data, and E. Kodytková for 9. Valiente Moro C, Thioulouse J, Chauve C, Normand P, Zenner L. Bacterial taxa asso- or IgG seroconversion in paired serum manuscript review. ciated with the hematophagous mite Der- samples were observed for all patients manyssus gallinae detected by 16S rRNA except the grandfather (Table). Oto Melter, Mardjan Arvand, PCR amplifi cation and TTGE fi ngerprint- Oral clarithromycin and ing. Res Microbiol. 2009;160:63–70. Jiří Votýpka, doi:10.1016/j.resmic.2008.10.006 doxycycline were administered to and Dagmar Hulínská 10. Reeves WK, Dowling APG, Dasch GA. the children and adults, respectively, Author affi liations: Charles University, Rickettsial agents from parasitic Derma- nyssoidea (Acari: Mesostigmata). Exp for 10 days. The apartment was Prague, Czech Republic (O. Melter); repeatedly treated with insecticide, Appl Acarol. 2006;38:181–8. doi:10.1007/ Zentrum für Gesundheitsschutz, Dillenburg, s10493-006-0007-1 and the hole in the roof was repaired, Germany (M. Arvand); and National Institute leading to eradication of the mites. -

Découpage Électoral
PROJET IBOGA Découpage électoral Edité le :jeudi 28 avril 2016 PROVINCE ESTUAIRE COMMUNE/DEPARTEMENT LIBREVILLE ARRONDISSEMENT/CANTON 1ER ARRONDISSEMENT CODE CENTRE DE VOTE INSCRITS BUREAUX 01 01 01 01 ECOLE PUBL. BAS DE GUE-GUE 1 158 3 01 01 01 02 ECOLE PUBLIQUE OKALA 4 531 10 01 01 01 03 C.E.S. ANGE MBA 19 1 01 01 01 04 LYCEE INDJENDJE GONDJOUT 862 2 01 01 01 05 LYCEE LEON MBA 1 920 4 01 01 01 06 ECOLE CONV. CHARBONNAGES 1 695 4 01 01 01 07 ECOLE PUBLIQUE AMBOWE 2 006 5 01 01 01 08 ECOLE PUBLIQUE ALIBADENG 1 958 4 01 01 01 09 ECOLE PUBLIQUE DE LOUIS 5 708 12 01 01 01 10 ECOLE CONV. GROS BOUQUET 1 822 2 01 01 01 11 ECOLE CONV. GROS BOUQUET 2 249 1 01 01 01 12 ECOLE PUBLIQUE BA OUMAR 771 2 01 01 01 13 ECOLE PUBL. -ENS- A 1 076 3 01 01 01 14 ECOLE PUBL. ENSET.B 230 1 01 01 01 15 CES PRIVE DE LOUIS 64 1 01 01 01 16 ECOLE PUBL. -ENS- B 667 2 01 01 01 17 ECOLE PUBLIQUE CHARBONNAGES 5 752 12 01 01 01 18 ECOLE PUBL. ENSET A 481 1 01 01 01 19 COLLEGE N.DAME DE QUABEN 1 015 3 01 01 01 20 EP. GROS BOUQUET 4 571 2 01 01 01 21 LYCEE NELSON MANDELA 3 514 8 01 01 01 22 EC-PUBL-OKALA B 46 1 TOTAL 35 115 84 Page 2 / 457 PROVINCE ESTUAIRE COMMUNE/DEPARTEMENT LIBREVILLE ARRONDISSEMENT/CANTON 2EME ARRONDISSEMENT CODE CENTRE DE VOTE INSCRITS BUREAUX 01 01 02 01 ECOLE ST MICHEL (B) 1 304 3 01 01 02 02 ECOLE PUBLIQUE SOTEGA 1 209 3 01 01 02 03 ECOLE SAINT JOSEPH 1 208 1 01 01 02 04 ECOLE PUBL. -

Etat Des Conna1ssances Sur Les Tortues Continentales Du Gabon: Distribution, Ecologie Et Conservation Par Jerome MARAN & Olivier S.G
II BULLETIN DE L'INSTITUT ROYAL DES SCIENCES NATUREL LES DE BELGIQUE, BIOLOGIE, 75 : 47-60, 2005 BULLETIN VAN HET KONINKLIJK BELGISCH INST IT UUT VOOR NATUURWETENSCHAPPEN, BIOLOGIE, 75 : 47-60, 2005 Etat des conna1ssances sur les tortues continentales du Gabon: distribution, ecologie et conservation par Jerome MARAN & Olivier S.G. PAUWELS Abstract Guinee-Equatoriale, au nord par Ia Republique du Came roun , a ]'est et au sud par Republique du Congo (ou Congo New observations made by th e authors over the whole territory of Brazzaville) eta !'ouest par !'Ocean atlantique, sur plus de Gabon all ow to strongly in crease th e knowledge on the di stributi on 750 km de cotes. Les forets denses humides sempervirentes of the local terrestrial and fre shwater chelon ians. The research et semi-sempervirentes couvrent 77, 2 % de Ia superficie to results bring also numerous new data on their natural hi sto ry, tale du Gabon. On distingue neuf grands types de fOimations morphol ogy, ecology and ex pl oit ation. All nine currentl y recorded vegetales, dont six appartiennent au domaine forestier (CA species (Pelomed usid ae: Pelusios carina/U s, P. castaneus, P. BALLE, 1983). I' ins tar des au tre s regions equ atori ales, les chapini, P. gabonensis, P. marani and P. niger; Testudinidae: A Kinixys erosa; Tri onyc hid ae: Cyclodemw aubryi and Trionyx groupes zoologiques presents au Gabon man trent une grande triunguis) suffer from a hi gh human predation pressure. Some po diversification. Neanm oi ns, le Gabon demeure du point de pul ati ons mi ght seriously dec rease in the coming decades, vue herpetologique l' un des pays d'Afrique centrale les es peciall y Trionyc hidae. -

Exposure to Ebola Virus and Risk for Infection with Malaria Parasites, Rural Gabon
Article DOI: https://doi.org/10.3201/eid2602.181120 Exposure to Ebola Virus and Risk for Infection with Malaria Parasites, Rural Gabon Appendix Supplementary Data and Analyses Population-level data Appendix Fig 1 Appendix Table 1 Department-level co-factor analysis Appendix Table 2 Appendix Fig 2 Appendix Fig 3 Malaria infection risk analysis Appendix Table 3 Appendix Fig 4 P. falciparum infection risk analysis Appendix Table 4 P. malariae infection risk analysis Appendix Table 5 Ebola virus exposure risk analysis Appendix Table 6 Appendix Fig 5 Appendix Fig 6 Page 1 of 13 Appendix Table 1. Population-level data. Most Common Population Population Average Wealth Index Frequency of ZEBOV- Province Number Number Size (2003 Density Household Quantile (% Households Malaria specific IgG (Date Villages Persons National (Persons per Wealth Score households) (DHS with an ITN Parasite Antibody Sampled) Department Sampled Sampled Census) km2) (DHS 2012) 2012) (DHS 2012) Prevalence Prevalence Estuaire (July 2005) KOMO 17 207 12690 1.28 -93740 poorest (87%) 0.72 72.5% 18.4% KOMO MONDAH 4 53 104302 28.58 8407 poorest (29.5%) 0.71 54.7% 7.5% NOYA 5 40 6268 1.58 -57261 poorest (76.9%) 0.94 65.0% 15.0% Haut-Ogooué (April 2007) DJOUE 1 18 3503 1.57 -119763 poorest (100%) 0.72 72.2% 5.6% DJOUORI AGNILI 1 24 4301 1.98 -81382 poorest (90%) 0.65 25.0% 4.2% LEBOMBI LEYOU 1 11 53921 20.74 -70923 poorest (86.7%) 0.65 18.2% 9.1% LEKABI LEWOLO 2 48 6417 5.77 -61302 poorest (80%) 0.68 66.7% 12.5% LEKOKO 1 14 3412 0.87 100.0% 42.9% LEKONI LEKORI 1 18 8978 6.79 61.1% -

Situation Socio-Économique - Ogooué - Maritime 2012
1 SITUATION SOCIO-ÉCONOMIQUE - OGOOUÉ - MARITIME 2012 Ogooue Maritime 2013.indd 1 26/06/14 09:32 2 SITUATION SOCIO-ÉCONOMIQUE - OGOOUÉ - MARITIME 2012 Ogooue Maritime 2013.indd 2 26/06/14 09:32 AVANT-PROPOS 7 INTRODUCTION 9 SOMMAIRE Partie I : CONNAITRE LA PROVINCE 11 I.1. – LA TERRE ET LES HOMMES 13 I.1.1. – La situation géographique 13 I.1.2. – La géographie physique 13 I.1.3. – La géographie humaine 15 I.1.4. – Le découpage administratif 16 I.2. – L’HISTOIRE ET LA CULTURE 17 I.2.1. – L’histoire 17 I.2.2. – La Culture et les rites traditionnels 18 I.2.3. – Les grands groupes ethnolinguistiques 18 Partie II : INFORMATIONS SOCIALES 21 II.1. – LA SANTE 23 II.1.1. – Les infrastructures 23 II.1.2. – Le personnel de santé 24 II.1.3. – L’accès des populations aux soins de santé 24 II.2. – L’EDUCATION 25 II.2.1. – Les structures éducatives 25 II.2.2. – Les effectifs des élèves et des enseignants 26 II.3. – LA FORMATION PROFESSIONNELLE 28 II.3.1. – Le Centre de Formation et de Perfectionnement Professionnel 28 II.3.2. – Les établissements techniques professionnels 30 3 SITUATION SOCIO-ÉCONOMIQUE - OGOOUÉ - MARITIME 2012 Ogooue Maritime 2013.indd 3 26/06/14 09:32 II.3.3. – Les structures privées de formation professionnelle 31 II.4. – L’EMPLOI 33 II.4.1. – L’emploi dans le secteur privé 33 SOMMAIRE II.4.2. – L’emploi dans les administrations publiques 34 II.4.3. – L’emploi des administrations décentralisées 35 II.5. -

Exposure to Ebola Virus and Risk for Infection with Malaria Parasites, Rural Gabon Jessica L
Exposure to Ebola Virus and Risk for Infection with Malaria Parasites, Rural Gabon Jessica L. Abbate, Pierre Becquart, Eric Leroy, Vanessa O. Ezenwa,1 Benjamin Roche1 diseases (1) and the high frequency of Plasmodium An association between malaria and risk for death spp. co-infection among patients undergoing treat- among patients with Ebola virus disease has suggested ment for confirmed EVD 2( ). At the individual level, within-host interactions between Plasmodium falciparum parasites and Ebola virus. To determine whether such several retrospective epidemiology studies of pa- an interaction might also influence the probability of ac- tients undergoing treatment for confirmed EVD have quiring either infection, we used a large snapshot sur- attempted to determine whether concurrent malaria veillance study from rural Gabon to test if past exposure affects patient outcomes. In Sierra Leone (3) and at to Ebola virus is associated with current infection with 1 Ebola treatment center in Liberia (4), mortality risk Plasmodium spp. during nonepidemic conditions. We was much higher among Ebola patients who were co- found a strong positive association, on population and infected with Plasmodium parasites than among pa- individual levels, between seropositivity for antibodies tients who were not co-infected, and a study in Guin- against Ebola virus and the presence of Plasmodium ea found that adverse outcomes were higher among parasites in the blood. According to a multiple regres- EVD patients with higher P. falciparum parasite loads sion model accounting for other key variables, antibodies than among those with lower levels of parasitemia (5). against Ebola virus emerged as the strongest individual- level risk factor for acquiring malaria.