De Novo Gonad Transcriptome Analysis of the Common Littoral Shrimp
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Systematic and Experimental Analysis of Their Genes, Genomes, Mrnas and Proteins; and Perspective to Next Generation Sequencing
Crustaceana 92 (10) 1169-1205 CRUSTACEAN VITELLOGENIN: A SYSTEMATIC AND EXPERIMENTAL ANALYSIS OF THEIR GENES, GENOMES, MRNAS AND PROTEINS; AND PERSPECTIVE TO NEXT GENERATION SEQUENCING BY STEPHANIE JIMENEZ-GUTIERREZ1), CRISTIAN E. CADENA-CABALLERO2), CARLOS BARRIOS-HERNANDEZ3), RAUL PEREZ-GONZALEZ1), FRANCISCO MARTINEZ-PEREZ2,3) and LAURA R. JIMENEZ-GUTIERREZ1,5) 1) Sea Science Faculty, Sinaloa Autonomous University, Mazatlan, Sinaloa, 82000, Mexico 2) Coelomate Genomic Laboratory, Microbiology and Genetics Group, Industrial University of Santander, Bucaramanga, 680007, Colombia 3) Advanced Computing and a Large Scale Group, Industrial University of Santander, Bucaramanga, 680007, Colombia 4) Catedra-CONACYT, National Council for Science and Technology, CDMX, 03940, Mexico ABSTRACT Crustacean vitellogenesis is a process that involves Vitellin, produced via endoproteolysis of its precursor, which is designated as Vitellogenin (Vtg). The Vtg gene, mRNA and protein regulation involve several environmental factors and physiological processes, including gonadal maturation and moult stages, among others. Once the Vtg gene, mRNAs and protein are obtained, it is possible to establish the relationship between the elements that participate in their regulation, which could either be species-specific, or tissue-specific. This work is a systematic analysis that compares the similarities and differences of Vtg genes, mRNA and Vtg between the crustacean species reported in databases with respect to that obtained from the transcriptome of Callinectes arcuatus, C. toxotes, Penaeus stylirostris and P. vannamei obtained with MiSeq sequencing technology from Illumina. Those analyses confirm that the Vtg obtained from selected species will serve to understand the process of vitellogenesis in crustaceans that is important for fisheries and aquaculture. RESUMEN La vitelogénesis de los crustáceos es un proceso que involucra la vitelina, producida a través de la endoproteólisis de su precursor llamado Vitelogenina (Vtg). -

Redalyc.Crecimiento Del Camarón Excavador Parastacus Pugnax
Latin American Journal of Aquatic Research E-ISSN: 0718-560X [email protected] Pontificia Universidad Católica de Valparaíso Chile Ibarra, Mauricio; Arana, Patricio M. Crecimiento del camarón excavador Parastacus pugnax (Poeppig, 1835) determinado mediante técnica de marcaje Latin American Journal of Aquatic Research, vol. 39, núm. 2, julio, 2011, pp. 378-384 Pontificia Universidad Católica de Valparaíso Valparaiso, Chile Disponible en: http://www.redalyc.org/articulo.oa?id=175019398018 Cómo citar el artículo Número completo Sistema de Información Científica Más información del artículo Red de Revistas Científicas de América Latina, el Caribe, España y Portugal Página de la revista en redalyc.org Proyecto académico sin fines de lucro, desarrollado bajo la iniciativa de acceso abierto Lat. Am. J. Aquat. Res., 39(2): 378-384, 2011 Lat. Am. J. Aquat. Res. 378 DOI: 10.3856/vol39-issue2-fulltext-18 Short Communication Crecimiento del camarón excavador Parastacus pugnax (Poeppig, 1835) determinado mediante técnica de marcaje Mauricio Ibarra1 & Patricio M. Arana1 1Escuela de Ciencias del Mar, Pontificia Universidad Católica de Valparaíso P.O. Box 1020, Valparaíso, Chile RESUMEN. Para determinar el crecimiento en el camarón excavador (Parastacus pugnax) en la zona centro sur de Chile se utilizó un marbete tipo cinturón. Los parámetros longitud cefalotorácica asintótica (Lc∞) y la velocidad de incremento en longitud y peso (K), se establecieron mediante el método de Gulland & Holt (1959). El parámetro t0 se determinó mediante la ecuación inversa del modelo de von Bertalanffy, estableciéndose que las curvas de crecimiento en longitud y peso fueron definidas por los parámetros K = 0,35 -1 mm año , t0 = -0,38 años, Lc∞ = 55,9 mm y W∞ = 83,8 g, valores similares a los de Samastacus spinifrons, especie chilena de alto potencial de cultivo, y de otros parastácidos sudamericanos, tales como P. -

Full Text in Pdf Format
Vol. 25: 83–96, 2016 AQUATIC BIOLOGY Published August 24 doi: 10.3354/ab00663 Aquat Biol OPENPEN ACCESSCCESS Thermal adaptations of embryos of six terrestrial hermit crab species Katsuyuki Hamasaki1,*, Takahiro Matsuda1, Ken Takano1, Mio Sugizaki1, Yu Murakami1, Shigeki Dan2, Shuichi Kitada1 1Graduate School of Marine Science and Technology, Tokyo University of Marine Science and Technology, Minato, Konan, Tokyo 108-8477, Japan 2Research Centre for Marine Invertebrate, National Research Institute of Fisheries and Environment of Inland Sea, Fisheries Research Agency, Momoshima, Onomichi, Hiroshima 722-0061, Japan ABSTRACT: We evaluated the thermal adaptations of embryos of 6 terrestrial hermit crab species in the family Coenobitidae (genera Birgus and Coenobita): B. latro, C. brevimanus, C. cavipes, C. purpureus, C. rugosus, and C. violascens. Embryos of each species were cultured in vitro at 6 dif- ferent temperatures (18 to 34°C) in artificial seawater to avoid air desiccation; the lower threshold temperatures for embryonic development were estimated using heat summation theory equa- tions. Additionally, partial effective cumulative temperatures (> lower threshold temperature) until hatching were determined for ovigerous females of each species cultured in containers. The relationships between the embryonic growth index values (relative area of the embryonic body vs. total embryo surface) and effective cumulative temperatures were expressed using cubic equa- tions. Lower threshold temperature was estimated to be 12.7 to 14.5°C. The effective cumulative temperature and egg incubation period estimates from the appearance of the embryonic body to hatching were higher in B. latro and C. brevimanus, followed by C. rugosus, C. cavipes, and C. violascens, and lower in C. -

Nutritional Composition, Antioxidants and Antimicrobial Activities In
Research Journal of Biotechnology Vol. 15 (4) April (2020) Res. J. Biotech Nutritional Composition, Antioxidants and Antimicrobial Activities in Muscle Tissues of Mud Crab, Scylla paramamosain Wan Roslina Wan Yusof1*, Noorasmin Mokhtar Ahmad2, Mohd Alhafiizh Zailani1, Mardhiah Mohd Shahabuddin1, Ngieng Ngui Sing2 and Awang Ahmad Sallehin Awang Husaini2 1. Centre for Pre-University Studies, Universiti Malaysia Sarawak, 94300 Kota Samarahan, Sarawak, MALAYSIA 2. Faculty of Resource Science and Technology, Universiti Malaysia Sarawak, 94300 Kota Samarahan, Sarawak, MALAYSIA *[email protected] Abstract paramamosain, Scylla serrata, Scylla transquebarica and Mud crab, Scylla paramamosain known as a green mud Scylla olivacea. Interestingly, S. paramamosain also known crab, has become a popular seafood due to its meat as a green mud crab is one of the popular species and widely quality. In addition, this marine invertebrate has been distributed in mangrove area with high water salinity such as continental coast of South China Sea and Java Sea.10 found to possess peptides with different biological activities and potentials. The aim was, first, to Among the other species, S. paramamosain has triangular determine the basic nutritional content and second, to frontal lobe spines and easily identified by the dotted pattern screen for the antioxidants and antimicrobials on its propodus. From nutritional point of view, mud crabs activities in the tissue of mud crab, S. paramamosain. have high protein, minerals and polyunsaturated fatty acids Percentages of carbohydrate, protein and fat in S. contents.23 Apart from nutritional view, many studies have paramamosain were 2.32%, 12.53% and 0.23% been conducted on the biological activities in mud crabs, respectively. -

Feeding Habits of the Prawns Processa Edulzs and Palaemon Adspersus (Crustacea, Decapoda, Caridea) in the Alfacs Bay, Ebro Delta (Nw Mediterranean)
FEEDING HABITS OF THE PRAWNS PROCESSA EDULZS AND PALAEMON ADSPERSUS (CRUSTACEA, DECAPODA, CARIDEA) IN THE ALFACS BAY, EBRO DELTA (NW MEDITERRANEAN) Guerao, G., 1993-1994. Feeding habits of the prawns Processa edulis and Palaemon adspersus (Crustacea, Decapoda, Caridea) in the Alfacs Bay, Ebro Delta (NW Mediterranean). Misc. Zool., 17: 115-122. Feeding habits of the prawns Processa edulis and Palaemon adspersus (Crustacea, Decapoda, Caridea) in the Alfacs Bay, Ebro Delta (NW Mediterranean).- The stomach contents of 147 Palaemon adspersus Rathke, and 102 Processa edulis (Risso) were analyzed. The frequency of occurrence method and the points method were used. The role of these species in the food web of Cymodocea nodosa meadows is defined. Results indicate that both species are predators of benthic invertebrates rather than scavengers or detritus feeders. The main food items varied according to species. The diet of Palaemon adspersus consisted almost entirely of crustaceans, molluscs, and plant material, with amphipods playing a major role. Processa edulis ate an almost equal amount of crustaceans and polychaetes. In P. adspersus, most dietary items differed according to size classes of prawn. Key words: Feeding, Prawns, Palaemon, Processa, Ebro Delta. (Rebut: 18 V 94; Acceptació condicional: 13 IX 94; Acc. definitiva: 18 X 94) G. Guerao, Dept. de Biologia Animal (Artrdpodes), Fac. de Biologia, Univ. de Barcelona, Avgda. Diagonal 645, 08028 Barcelona, Espanya (Spain). INTRODUCTION and has been recorded from as far north as the Norwegian Sea to the Marocco coast Processidae prawns are abundant in coastal (LAGARDERE,1971) and the Mediterranean waters of temperate and tropical areas. (ZARIQUIEYÁLVAREZ, 1968). This species is Processa edulis (Risso, 1816) is a comrnon the subject of commercial fisheries in many littoral mediterranean prawn (ZARIQUIEY areas (JENSEN,1958; HOLTHUIS,1980; ÁLVAREZ,1968). -

Homarus Gammarus ) Reveals Its Generalist Parasitic Strategy in Marine Invertebrates
Title Halioticida noduliformans infection in eggs of lobster ( Homarus gammarus ) reveals its generalist parasitic strategy in marine invertebrates Authors Holt, C; Foster, R; Daniels, CL; van der Giezen, M; Feist, SW; Stentiford, GD; Bass, D Description publisher: Elsevier articletitle: Halioticida noduliformans infection in eggs of lobster (Homarus gammarus) reveals its generalist parasitic strategy in marine invertebrates journaltitle: Journal of Invertebrate Pathology articlelink: http://dx.doi.org/10.1016/ j.jip.2018.03.002 content_type: article copyright: © 2018 The Authors. Published by Elsevier Inc.; 0000-0002-6719-5565 Date Submitted 2018-06-02 Journal of Invertebrate Pathology xxx (xxxx) xxx–xxx Contents lists available at ScienceDirect Journal of Invertebrate Pathology journal homepage: www.elsevier.com/locate/jip Halioticida noduliformans infection in eggs of lobster (Homarus gammarus) reveals its generalist parasitic strategy in marine invertebrates ⁎ Corey Holta,b,c, , Rachel Fosterd, Carly L. Danielsc, Mark van der Giezenb, Stephen W. Feista, Grant D. Stentiforda, David Bassa,d a Pathology and Microbial Systematics, Centre for Environment, Fisheries and Aquaculture Science (Cefas), Barrack Road, Weymouth, Dorset DT4 8UB, United Kingdom b Biosciences, University of Exeter, Stocker Road, Exeter EX4 4QD, United Kingdom c The National Lobster Hatchery, South Quay, Padstow PL28 9BL, United Kingdom d The Natural History Museum, Cromwell Road, Kensington, London SW7 5BD, United Kingdom ARTICLE INFO ABSTRACT Keywords: A parasite exhibiting Oomycete-like morphology and pathogenesis was isolated from discoloured eggs of the Halioticida noduliformans European lobster (Homarus gammarus) and later found in gill tissues of adults. Group-specific Oomycete primers Homarus gammarus were designed to amplify the 18S ribosomal small subunit (SSU), which initially identified the organism as the Haliphthoros same as the ‘Haliphthoros’ sp. -

Full Text in Pdf Format
MARINE ECOLOGY PROGRESS SERIES Published November 14 Mar Ecol Prog Ser Long-term distribution patterns of mobile estuarine invertebrates (Ctenophora, Cnidaria, Crustacea: Decapoda) in relation to hydrological parameters Martin J. Attrill'.', R. Myles ~homas~ 'Marine Biology & Ecotoxicology Research Group, Plymouth Environmental Research Centre, University of Plymouth, Drake Circus, Plymouth PL4 8.4.4, United Kingdom 'Environment Agency (Thames Region), Aspen House, Crossbrook St., Waltham Cross, Herts EN8 8HE, United Kingdom ABSTRACT: Between 1977 and 1992, semi-quantitative samples of macroinvertebrates were taken at fortnightly intervals from the Thames Estuary (UK) utilising the cooling water intake screens of West Thurrock power station. Samples were taken for 4 h over low water, the abundances of invertebrates recorded in 30 min subsamples and related to water volume filtered. Abundances of the major estu- arine species have therefore been recorded every 2 wk for a 16 yr period, together wlth physico- chemical parameters such as temperature, salinity and freshwater flow. Annual cycles of distribution were apparent for several species. Carcinus rnaenas exhibited a regular annual cycle, with a peak in autumn followed by a decrease in numbers over winter, relating to seasonal temperature patterns. Conversely, abundance of Crangon crangon was consistently lowest in summer, responding to seasonal changes in salinity, whilst Liocarcinus holsatus, Aurelia aurita and Pleurobrachia pileus were only pre- sent in summer samples, with P. pileus often in vast numbers (>l00000 per 500 million I). The estuar- ine prawn Palaemon longirostris showed no obvious sustained annual pattern, but evidence for a longer cycle of distribution was apparent. During 1989-1992 severe droughts in southeast England severely disrupted annual salinity patterns and coincided with a large increase in the Chinese mitten crab Eriocheir sinensis population. -

Mud Crab Susceptibility to Disease from White Spot Syndrome Virus Is Species-Dependent Somboonna Et Al
Mud crab susceptibility to disease from white spot syndrome virus is species-dependent Somboonna et al. Somboonna et al. BMC Research Notes 2010, 3:315 http://www.biomedcentral.com/1756-0500/3/315 (20 November 2010) Somboonna et al. BMC Research Notes 2010, 3:315 http://www.biomedcentral.com/1756-0500/3/315 SHORT REPORT Open Access Mud crab susceptibility to disease from white spot syndrome virus is species-dependent Naraporn Somboonna1,2,5*, Seksan Mangkalanan3, Attasit Udompetcharaporn2, Chartchai Krittanai3, Kallaya Sritunyalucksana1,2, TW Flegel1,2,4 Abstract Background: Based on a report for one species (Scylla serrata), it is widely believed that mud crabs are relatively resistant to disease caused by white spot syndrome virus (WSSV). We tested this hypothesis by determining the degree of susceptibility in two species of mud crabs, Scylla olivacea and Scylla paramamosain, both of which were identified by mitochondrial 16 S ribosomal gene analysis. We compared single-dose and serial-dose WSSV challenges on S. olivacea and S. paramamosain. Findings: In a preliminary test using S. olivacea alone, a dose of 1 × 106 WSSV copies/g gave 100% mortality within 7 days. In a subsequent test, 17 S. olivacea and 13 S. paramamosain were divided into test and control groups for challenge with WSSV at 5 incremental, biweekly doses starting from 1 × 104 and ending at 5 × 106 copies/g. For 11 S. olivacea challenged, 3 specimens died at doses between 1 × 105 and 5 × 105 copies/g and none died for 2 weeks after the subsequent dose (1 × 106 copies/g) that was lethal within 7 days in the preliminary test. -

Palaemon Serratus Fishery: Biology, Ecology & Management
BANGOR UNIVERSITY, FISHERIES AND CONSERVATION REPORT NO.38 A review of the Palaemon serratus fishery: biology, ecology & management Haig, J., Ryan, N.M., Williams, K.F. & M.J. Kaiser June 2014 Please cite as follows: Haig, J., Ryan, N.M., Williams, K.F. & M.J. Kaiser (2014) A review of the Palaemon serratus fishery: biology, ecology & management. Bangor University, Fisheries and Conservation Report No. 38. Funded By: Bangor University, Fisheries and Conservation Report No. 38 PURPOSE This paper reviews the biology, ecology and management of the commercially fished shrimp species Palaemon serratus. The purpose of the review is to identify gaps in current knowledge so that we can identify the required research that will assist in assessing and managing Palaemon stocks to ensure their future sustainability. INTRODUCTION Palaemon serratus is a decapod crustacean that inhabits inshore coastal areas around the majority of the coast of the UK and Ireland. It is a relatively short-lived species with a life span of between two (Forster 1951) and five years (Cole 1958). P. serratus can be found in either shallow intertidal pools or deeper subtidal waters. They appear to be migratory; occuring in high abundances in shallow waters during summer months and in deeper water (up to 40m) during the winter. P. serratus has a number of historical synonymised names. It was first described by Pennant in 1777 as Astacus serratus and has since undergone several genera and species name changes up until 1950 (MarLIN & WoRMS, 2014). P. serratus is similar in morphology to other Palaemonid shrimp in northern temperate waters (e.g. -

The Crustacean Society Mid-Year Meeting 2019
THE CRUSTACEAN SOCIETY MID-YEAR MEETING 2019 ABSTRACT BOOKLET Table of Contents PLENARY LECTURES ........................................................................................................... 1 ORAL PRESENTATIONS ...................................................................................................... 7 SYMPOSIUM 1: Frontiers in Crustacean Biology: Asian Perspectives ................................ 43 SYMPOSIUM 2: Recent Advances in Caridean Systematics ............................................... 53 SYMPOSIUM 3: Evolution and Ecology of Parasitic and Symbiotic Crustaceans ................ 59 SYMPOSIUM 4: Biology of Freshwater Crayfish ................................................................ 69 SYMPOSIUM 5: Deep-sea Biodiversity: A Crustacean Perspective .................................... 77 SYMPOSIUM 6: Comparative Endocrinology and Genomics in Arthropods ....................... 87 SYMPOSIUM 7: Fossil and Modern Clam Shrimp .............................................................. 97 SYMPOSIUM 8: Aquaculture Biotechnology of Crabs ..................................................... 108 POSTER PRESENTATIONS ............................................................................................... 114 PLENARY LECTURES PL1 Effects of temperature variations on reproduction: Transduction of physiological stress through species interactions between two porcelain crabs B. TSUKIMURA1, ALEX GUNDERSON2, JONATHON STILLMAN3 1. California State University, Fresno, USA 2. Tulane University, USA 3. -

Larval Growth
LARVAL GROWTH Edited by ADRIAN M.WENNER University of California, Santa Barbara OFFPRINT A.A.BALKEMA/ROTTERDAM/BOSTON DARRYL L.FELDER* / JOEL W.MARTIN** / JOSEPH W.GOY* * Department of Biology, University of Louisiana, Lafayette, USA ** Department of Biological Science, Florida State University, Tallahassee, USA PATTERNS IN EARLY POSTLARVAL DEVELOPMENT OF DECAPODS ABSTRACT Early postlarval stages may differ from larval and adult phases of the life cycle in such characteristics as body size, morphology, molting frequency, growth rate, nutrient require ments, behavior, and habitat. Primarily by way of recent studies, information on these quaUties in early postlarvae has begun to accrue, information which has not been previously summarized. The change in form (metamorphosis) that occurs between larval and postlarval life is pronounced in some decapod groups but subtle in others. However, in almost all the Deca- poda, some ontogenetic changes in locomotion, feeding, and habitat coincide with meta morphosis and early postlarval growth. The postmetamorphic (first postlarval) stage, here in termed the decapodid, is often a particularly modified transitional stage; terms such as glaucothoe, puerulus, and megalopa have been applied to it. The postlarval stages that fol low the decapodid successively approach more closely the adult form. Morphogenesis of skeletal and other superficial features is particularly apparent at each molt, but histogenesis and organogenesis in early postlarvae is appreciable within intermolt periods. Except for the development of primary and secondary sexual organs, postmetamorphic change in internal anatomy is most pronounced in the first several postlarval instars, with the degree of anatomical reorganization and development decreasing in each of the later juvenile molts. -
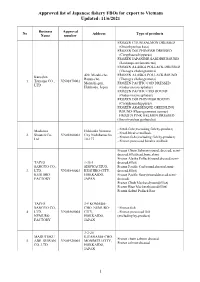
Approved List of Japanese Fishery Fbos for Export to Vietnam Updated: 11/6/2021
Approved list of Japanese fishery FBOs for export to Vietnam Updated: 11/6/2021 Business Approval No Address Type of products Name number FROZEN CHUM SALMON DRESSED (Oncorhynchus keta) FROZEN DOLPHINFISH DRESSED (Coryphaena hippurus) FROZEN JAPANESE SARDINE ROUND (Sardinops melanostictus) FROZEN ALASKA POLLACK DRESSED (Theragra chalcogramma) 420, Misaki-cho, FROZEN ALASKA POLLACK ROUND Kaneshin Rausu-cho, (Theragra chalcogramma) 1. Tsuyama CO., VN01870001 Menashi-gun, FROZEN PACIFIC COD DRESSED LTD Hokkaido, Japan (Gadus macrocephalus) FROZEN PACIFIC COD ROUND (Gadus macrocephalus) FROZEN DOLPHIN FISH ROUND (Coryphaena hippurus) FROZEN ARABESQUE GREENLING ROUND (Pleurogrammus azonus) FROZEN PINK SALMON DRESSED (Oncorhynchus gorbuscha) - Fresh fish (excluding fish by-product) Maekawa Hokkaido Nemuro - Fresh bivalve mollusk. 2. Shouten Co., VN01860002 City Nishihamacho - Frozen fish (excluding fish by-product) Ltd 10-177 - Frozen processed bivalve mollusk Frozen Chum Salmon (round, dressed, semi- dressed,fillet,head,bone,skin) Frozen Alaska Pollack(round,dressed,semi- TAIYO 1-35-1 dressed,fillet) SANGYO CO., SHOWACHUO, Frozen Pacific Cod(round,dressed,semi- 3. LTD. VN01840003 KUSHIRO-CITY, dressed,fillet) KUSHIRO HOKKAIDO, Frozen Pacific Saury(round,dressed,semi- FACTORY JAPAN dressed) Frozen Chub Mackerel(round,fillet) Frozen Blue Mackerel(round,fillet) Frozen Salted Pollack Roe TAIYO 3-9 KOMABA- SANGYO CO., CHO, NEMURO- - Frozen fish 4. LTD. VN01860004 CITY, - Frozen processed fish NEMURO HOKKAIDO, (excluding by-product) FACTORY JAPAN