Zebrafish Staufen1 and Staufen2 Are Required for the Survival And
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

STAU2 (NM 001164380) Human Untagged Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for SC327098 STAU2 (NM_001164380) Human Untagged Clone Product data: Product Type: Expression Plasmids Product Name: STAU2 (NM_001164380) Human Untagged Clone Tag: Tag Free Symbol: STAU2 Synonyms: 39K2; 39K3 Vector: pCMV6-Entry (PS100001) E. coli Selection: Kanamycin (25 ug/mL) Cell Selection: Neomycin This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 3 STAU2 (NM_001164380) Human Untagged Clone – SC327098 Fully Sequenced ORF: >NCBI ORF sequence for NM_001164380, the custom clone sequence may differ by one or more nucleotides ATGGCAAACCCAAAAGAGAAAACTGCAATGTGTCTGGTAAATGAGTTAGCCCGTTTCAATAGAGTCCAAC CCCAGTATAAACTTCTGAATGAAAGAGGGCCTGCTCATTCAAAGATGTTCTCAGTGCAGCTGAGTCTTGG TGAGCAGACATGGGAATCCGAAGGCAGCAGTATAAAGAAGGCTCAGCAGGCTGTTGCCAATAAAGCTTTG ACTGAATCTACGCTTCCCAAACCAGTTCAGAAGCCACCCAAAAGTAATGTTAACAATAACCCAGGCAGTA TAACTCCAACTGTGGAACTGAATGGGCTTGCTATGAAAAGGGGAGAGCCTGCCATCTACAGGCCATTAGA TCCAAAGCCATTCCCAAATTATAGAGCTAATTACAACTTTCGGGGCATGTACAATCAGAGGTATCATTGC CCAGTGCCTAAGATCTTTTATGTTCAGCTCACTGTAGGAAATAATGAATTTTTTGGGGAAGGAAAGACTC GACAAGCTGCTAGACACAATGCTGCAATGAAAGCCCTCCAAGCACTGCAGAATGAACCTATTCCAGAAAG ATCTCCTCAGAATGGTGAATCAGGAAAGGATGTGGATGATGACAAAGATGCAAATAAGTCTGAGATCAGC TTAGTGTTTGAAATTGCTCTGAAGCGAAATATGCCTGTCAGTTTTGAGGTTATTAAAGAAAGTGGACCAC -

Anti-STAU2 Monoclonal Antibody, Clone 2D7 (DCABH-6925) This Product Is for Research Use Only and Is Not Intended for Diagnostic Use
Anti-STAU2 monoclonal antibody, clone 2D7 (DCABH-6925) This product is for research use only and is not intended for diagnostic use. PRODUCT INFORMATION Product Overview Mouse monoclonal to STAU2 - C-terminal Antigen Description RNA-binding protein required for the microtubule-dependent transport of neuronal RNA from the cell body to the dendrite. As protein synthesis occurs within the dendrite, the localization of specific mRNAs to dendrites may be a prerequisite for neurite outgrowth and plasticity at sites distant from the cell body. Immunogen Synthetic peptide within Human STAU2 aa 379-570 (C terminal). The exact sequence is proprietary.Database link: Q9NUL3 Source/Host Mouse Species Reactivity Mouse, Rat, Human Clone 2D7 Conjugate Unconjugated Applications WB, ICC/IF Format Liquid Size 125 μl Buffer pH: 9.0; Purification elution buffer is: 0.1 M glycine-HCl, pH 2.7 + Neutralizing buffer: 1 M Tris- HCl Preservative None Storage Store at +4°C short term (1-2 weeks). Upon delivery aliquot. Store at -20°C long term. Avoid freeze / thaw cycle. Ship Shipped at 4°C. GENE INFORMATION Gene Name STAU2 staufen, RNA binding protein, homolog 2 (Drosophila) [ Homo sapiens ] 45-1 Ramsey Road, Shirley, NY 11967, USA Email: [email protected] Tel: 1-631-624-4882 Fax: 1-631-938-8221 1 © Creative Diagnostics All Rights Reserved Official Symbol STAU2 Synonyms STAU2; staufen, RNA binding protein, homolog 2 (Drosophila); staufen (Drosophila, RNA binding protein) homolog 2; double-stranded RNA-binding protein Staufen homolog 2; 39K2; staufen homolog 2; 39K3; MGC119606; DKFZp781K0371; Entrez Gene ID 27067 Protein Refseq NP_001157852 UniProt ID Q9NUL3 Chromosome Location 8q21.11 Function double-stranded RNA binding; 45-1 Ramsey Road, Shirley, NY 11967, USA Email: [email protected] Tel: 1-631-624-4882 Fax: 1-631-938-8221 2 © Creative Diagnostics All Rights Reserved. -

Novel Extensions of Label Propagation for Biomarker Discovery in Genomic Data
NOVEL EXTENSIONS OF LABEL PROPAGATION FOR BIOMARKER DISCOVERY IN GENOMIC DATA by Matthew E. Stokes B.S. Systems and Control Engineering, Case Western Reserve University, 2008 M.S. Intelligent Systems Program / Biomedical Informatics, University of Pittsburgh 2011 Submitted to the Graduate Faculty of the Kenneth P. Dietrich School of Arts and Sciences in fulfillment of the requirements for the degree of Doctor of Philosophy University of Pittsburgh 2014 UNIVERSITY OF PITTSBURGH DIETRICH SCHOOL OF ARTS AND SCIENCES This dissertation proposal was presented by Matthew E. Stokes on July 17, 2014 and approved by M. Michael Barmada, PhD, Department of Human Genetics Gregory F. Cooper, MD, PhD, Department of Biomedical Informatics and the Intelligent Systems Program Milos Hauskrecht, PhD, Department of Computer Science and the Intelligent Systems Program Dissertation Advisor: Shyam Visweswaran, MD, PhD, Department of Biomedical Informatics and the Intelligent Systems Program ii NOVEL EXTENSIONS OF LABEL PORPAGATION FOR BIOMARKER DISCOVERY IN GENOMIC DATA Matthew E. Stokes, M.S University of Pittsburgh, 2014 Copyright © by Matthew E. Stokes 2014 iii NOVEL EXTENSIONS OF LABEL PROPAGATION FOR BIOMARKER DISCOVERY IN GENOMIC DATA Matthew E. Stokes, PhD University of Pittsburgh, 2014 One primary goal of analyzing genomic data is the identification of biomarkers which may be causative of, correlated with, or otherwise biologically relevant to disease phenotypes. In this work, I implement and extend a multivariate feature ranking algorithm called label propagation (LP) for biomarker discovery in genome-wide single-nucleotide polymorphism (SNP) data. This graph-based algorithm utilizes an iterative propagation method to efficiently compute the strength of association between a SNP and a phenotype. -

BMC Biology Biomed Central
BMC Biology BioMed Central Research article Open Access Classification and nomenclature of all human homeobox genes PeterWHHolland*†1, H Anne F Booth†1 and Elspeth A Bruford2 Address: 1Department of Zoology, University of Oxford, South Parks Road, Oxford, OX1 3PS, UK and 2HUGO Gene Nomenclature Committee, European Bioinformatics Institute (EMBL-EBI), Wellcome Trust Genome Campus, Hinxton, Cambridgeshire, CB10 1SA, UK Email: Peter WH Holland* - [email protected]; H Anne F Booth - [email protected]; Elspeth A Bruford - [email protected] * Corresponding author †Equal contributors Published: 26 October 2007 Received: 30 March 2007 Accepted: 26 October 2007 BMC Biology 2007, 5:47 doi:10.1186/1741-7007-5-47 This article is available from: http://www.biomedcentral.com/1741-7007/5/47 © 2007 Holland et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: The homeobox genes are a large and diverse group of genes, many of which play important roles in the embryonic development of animals. Increasingly, homeobox genes are being compared between genomes in an attempt to understand the evolution of animal development. Despite their importance, the full diversity of human homeobox genes has not previously been described. Results: We have identified all homeobox genes and pseudogenes in the euchromatic regions of the human genome, finding many unannotated, incorrectly annotated, unnamed, misnamed or misclassified genes and pseudogenes. -

Evidence for Convergent Evolution of SINE-Directed Staufen-Mediated Mrna Decay
Evidence for convergent evolution of SINE-directed Staufen-mediated mRNA decay Bronwyn A. Lucasa,b, Eitan Lavic, Lily Shiued, Hana Choa,b, Sol Katzmane, Keita Miyoshia,b, Mikiko C. Siomif, Liran Carmelc, Manuel Ares Jr.d,1, and Lynne E. Maquata,b,1 aDepartment of Biochemistry and Biophysics, School of Medicine and Dentistry, University of Rochester Medical Center, Rochester, NY 14642; bCenter for RNA Biology, University of Rochester, Rochester, NY 14642; cDepartment of Genetics, Alexander Silberman Institute of Life Sciences, Faculty of Science, Hebrew University of Jerusalem, Jerusalem 91904, Israel; dCenter for Molecular Biology of RNA, Department of Molecular, Cell and Developmental Biology, University of California, Santa Cruz, CA 95064; eCenter for Biomolecular Science and Engineering, University of California, Santa Cruz, CA 95064; and fGraduate School of Science, University of Tokyo, Tokyo 113-0032, Japan Edited by Marlene Belfort, University at Albany, Albany, NY, and approved December 11, 2017 (received for review September 1, 2017) Primate-specific Alu short interspersed elements (SINEs) as well as from 7SL RNA, whereas B2 elements were derived from tRNA, rodent-specific B and ID (B/ID) SINEs can promote Staufen-mediated B4 elements from the fusion of a tRNAAla-derived ID element at decay (SMD) when present in mRNA 3′-untranslated regions (3′-UTRs). the 5′-end and a B1 element at the 3′-end, and ID elements from The transposable nature of SINEs, their presence in long noncoding tRNAAla or a neuronally expressed brain cytoplasmic (BC1) RNAs, their interactions with Staufen, and their rapid divergence in RNA (1, 15–17). Alu and B or ID (B/ID) SINE families emerged different evolutionary lineages suggest they could have generated after primates and rodents diverged ∼90 million years ago substantial modification of posttranscriptional gene-control networks (MYA), and have continuously amplified independently in the during mammalian evolution. -

Reverse Engineering of Modified Genes by Bayesian Network Analysis Defines Olecm Ular Determinants Critical to the Development of Glioblastoma Brian W
Florida International University FIU Digital Commons Robert Stempel College of Public Health & Social Environmental Health Sciences Work 5-30-2013 Reverse Engineering of Modified Genes by Bayesian Network Analysis Defines olecM ular Determinants Critical to the Development of Glioblastoma Brian W. Kunkle Department of Environmental Health Sciences, Florida International University Changwon Yoo Department of Biostatistics, Florida International University, [email protected] Deodutta Roy Department of Environmental Health Sciences, Florida International University, [email protected] Follow this and additional works at: https://digitalcommons.fiu.edu/eoh_fac Part of the Medicine and Health Sciences Commons Recommended Citation Kunkle BW, Yoo C, Roy D (2013) Reverse Engineering of Modified Genes by Bayesian Network Analysis Defines Molecular Determinants Critical to the Development of Glioblastoma. PLoS ONE 8(5): e64140. https://doi.org/10.1371/ journal.pone.0064140 This work is brought to you for free and open access by the Robert Stempel College of Public Health & Social Work at FIU Digital Commons. It has been accepted for inclusion in Environmental Health Sciences by an authorized administrator of FIU Digital Commons. For more information, please contact [email protected]. Reverse Engineering of Modified Genes by Bayesian Network Analysis Defines Molecular Determinants Critical to the Development of Glioblastoma Brian W. Kunkle1, Changwon Yoo2, Deodutta Roy1* 1 Department of Environmental and Occupational Health, Florida International University, Miami, Florida, United States of America, 2 Department of Biostatistics, Florida International University, Miami, Florida, United States of America Abstract In this study we have identified key genes that are critical in development of astrocytic tumors. Meta-analysis of microarray studies which compared normal tissue to astrocytoma revealed a set of 646 differentially expressed genes in the majority of astrocytoma. -

393LN V 393P 344SQ V 393P Probe Set Entrez Gene
393LN v 393P 344SQ v 393P Entrez fold fold probe set Gene Gene Symbol Gene cluster Gene Title p-value change p-value change chemokine (C-C motif) ligand 21b /// chemokine (C-C motif) ligand 21a /// chemokine (C-C motif) ligand 21c 1419426_s_at 18829 /// Ccl21b /// Ccl2 1 - up 393 LN only (leucine) 0.0047 9.199837 0.45212 6.847887 nuclear factor of activated T-cells, cytoplasmic, calcineurin- 1447085_s_at 18018 Nfatc1 1 - up 393 LN only dependent 1 0.009048 12.065 0.13718 4.81 RIKEN cDNA 1453647_at 78668 9530059J11Rik1 - up 393 LN only 9530059J11 gene 0.002208 5.482897 0.27642 3.45171 transient receptor potential cation channel, subfamily 1457164_at 277328 Trpa1 1 - up 393 LN only A, member 1 0.000111 9.180344 0.01771 3.048114 regulating synaptic membrane 1422809_at 116838 Rims2 1 - up 393 LN only exocytosis 2 0.001891 8.560424 0.13159 2.980501 glial cell line derived neurotrophic factor family receptor alpha 1433716_x_at 14586 Gfra2 1 - up 393 LN only 2 0.006868 30.88736 0.01066 2.811211 1446936_at --- --- 1 - up 393 LN only --- 0.007695 6.373955 0.11733 2.480287 zinc finger protein 1438742_at 320683 Zfp629 1 - up 393 LN only 629 0.002644 5.231855 0.38124 2.377016 phospholipase A2, 1426019_at 18786 Plaa 1 - up 393 LN only activating protein 0.008657 6.2364 0.12336 2.262117 1445314_at 14009 Etv1 1 - up 393 LN only ets variant gene 1 0.007224 3.643646 0.36434 2.01989 ciliary rootlet coiled- 1427338_at 230872 Crocc 1 - up 393 LN only coil, rootletin 0.002482 7.783242 0.49977 1.794171 expressed sequence 1436585_at 99463 BB182297 1 - up 393 -

Multiscale Genomic Analysis of The
University of Tennessee Health Science Center UTHSC Digital Commons Theses and Dissertations (ETD) College of Graduate Health Sciences 5-2009 Multiscale Genomic Analysis of the Corticolimbic System: Uncovering the Molecular and Anatomic Substrates of Anxiety-Related Behavior Khyobeni Mozhui University of Tennessee Health Science Center Follow this and additional works at: https://dc.uthsc.edu/dissertations Part of the Mental and Social Health Commons, Nervous System Commons, and the Neurosciences Commons Recommended Citation Mozhui, Khyobeni , "Multiscale Genomic Analysis of the Corticolimbic System: Uncovering the Molecular and Anatomic Substrates of Anxiety-Related Behavior" (2009). Theses and Dissertations (ETD). Paper 180. http://dx.doi.org/10.21007/etd.cghs.2009.0219. This Dissertation is brought to you for free and open access by the College of Graduate Health Sciences at UTHSC Digital Commons. It has been accepted for inclusion in Theses and Dissertations (ETD) by an authorized administrator of UTHSC Digital Commons. For more information, please contact [email protected]. Multiscale Genomic Analysis of the Corticolimbic System: Uncovering the Molecular and Anatomic Substrates of Anxiety-Related Behavior Document Type Dissertation Degree Name Doctor of Philosophy (PhD) Program Anatomy and Neurobiology Research Advisor Robert W. Williams, Ph.D. Committee John D. Boughter, Ph.D. Eldon E. Geisert, Ph.D. Kristin M. Hamre, Ph.D. Jeffery D. Steketee, Ph.D. DOI 10.21007/etd.cghs.2009.0219 This dissertation is available at UTHSC Digital -

Accelerating Functional Gene Discovery in Osteoarthritis
The Jackson Laboratory The Mouseion at the JAXlibrary Faculty Research 2021 Faculty Research 1-20-2021 Accelerating functional gene discovery in osteoarthritis. Natalie C Butterfield Katherine F Curry Julia Steinberg Hannah Dewhurst Davide Komla-Ebri See next page for additional authors Follow this and additional works at: https://mouseion.jax.org/stfb2021 Part of the Life Sciences Commons, and the Medicine and Health Sciences Commons Authors Natalie C Butterfield, Katherine F Curry, Julia Steinberg, Hannah Dewhurst, Davide Komla-Ebri, Naila S Mannan, Anne-Tounsia Adoum, Victoria D Leitch, John G Logan, Julian A Waung, Elena Ghirardello, Lorraine Southam, Scott E Youlten, J Mark Wilkinson, Elizabeth A McAninch, Valerie E Vancollie, Fiona Kussy, Jacqueline K White, Christopher J Lelliott, David J Adams, Richard Jacques, Antonio C Bianco, Alan Boyde, Eleftheria Zeggini, Peter I Croucher, Graham R Williams, and J H Duncan Bassett ARTICLE https://doi.org/10.1038/s41467-020-20761-5 OPEN Accelerating functional gene discovery in osteoarthritis Natalie C. Butterfield 1, Katherine F. Curry1, Julia Steinberg 2,3,4, Hannah Dewhurst1, Davide Komla-Ebri 1, Naila S. Mannan1, Anne-Tounsia Adoum1, Victoria D. Leitch 1, John G. Logan1, Julian A. Waung1, Elena Ghirardello1, Lorraine Southam2,3, Scott E. Youlten 5, J. Mark Wilkinson 6,7, Elizabeth A. McAninch 8, Valerie E. Vancollie 3, Fiona Kussy3, Jacqueline K. White3,9, Christopher J. Lelliott 3, David J. Adams 3, Richard Jacques 10, Antonio C. Bianco11, Alan Boyde 12, ✉ ✉ Eleftheria Zeggini 2,3, Peter I. Croucher 5, Graham R. Williams 1,13 & J. H. Duncan Bassett 1,13 1234567890():,; Osteoarthritis causes debilitating pain and disability, resulting in a considerable socio- economic burden, yet no drugs are available that prevent disease onset or progression. -
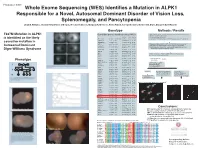
T237M Mutation in ALPK1 Is Identified As the Likely Causative Mutation In
Program #: 3367 Whole Exome Sequencing (WES) Identifies a Mutation in ALPK1 Responsible for a Novel, Autosomal Dominant Disorder of Vision Loss, Splenomegaly, and Pancytopenia Lloyd B. Williams, Chad D. Huff, Denise J. Morgan, Rosann Robinson, Margaux A. Morrison, Krista Kinard, George Rodgers, Kathleen B. Digre, Margaret M. DeAngelis Genotype Methods / Results WES on 4 individuals - 2 affected II-3 III-2, and 2 unaffected II-2 and III-3 T237M Mutation in ALPK1 Candi date genes identified using VAAST Illuimina Truseq Enrichment Kit position Allele Protein Quantitative PCR enrichment is identified as the likely Gene name p-value chromosome (hg19) change change Sequencing with Illumina HiSeq 2000 101 cycle paired end sequencing PRAMEF11 6.10E-06 chr1 12887174 C->T R->H causative mutation in ANKRD20A4 7.32E-06 chr9 69423637 G->A E->K MRPL4 8.55E-06 chr19 10367459 C->T R->W Mapped and aligned with Picard Tools (http://picard.sourceforge.net) FAM90A10 9.77E-06 chr8 7629232 G->T A->S SNPs and indels identified with Genome analysis toolkit (GATK) Autosomal Dominant Manual inspection and curation was done with Intergrative Genomics GOLGA6L10 9.77E-06 chr15 82635194 T->C E->G Viewer (http://www.broadinstitute.org/igv) Digre-Williams Syndrome FAM90A20 1.28E-05 chr8 7155458 C->G A->G EEF1A1 1.65E-05 chr6 74228474 C->T R->H MSI1 1.65E-05 chr12 120800875 C->T V->M VAAST - compares to HGMD, dbSNP, ESP, and1000 Genomes VDAC2 1.71E-05 chr10 76980685 G->T A->S panels to rule out common variants. PIM1 2.08E-05 chr6 37138779 A->T K->M USP11 2.26E-05 chrX -

Variants in STAU2 Associate with Metformin Response in a Type 2
medRxiv preprint doi: https://doi.org/10.1101/2020.03.18.20037218; this version posted March 20, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. It is made available under a CC-BY-NC-ND 4.0 International license . 1 Variants in STAU2 associate with metformin response in a type 2 2 diabetes cohort: a pharmacogenomics study using real-world 3 electronic health record data 4 5 Yanfei Zhang1, Ying Hu2*, Kevin Ho3*, Dustin N. Hartzel4, Vida Abedi5, Ramin Zand6, Marc S. Williams1, 6 Ming Ta M. Lee1 7 1. Genomic Medicine Institute, Geisinger, Danville, PA, USA 8 2. Department of endocrinology, Geisinger, Danville, PA, USA 9 3. Kidney Research Institute, Geisinger, Danville, PA, USA 10 4. Phenomic Analytics and Clinical Data Core, Geisinger, PA, USA 11 5. Department of molecular and functional genomics, Geisinger, Danville, PA, USA 12 6. Department of neurology, Neuroscience Institute, Geisinger, Danville, PA, USA 13 14 * Both Ying Hu (currently employed by Main Line Health) and Kevin Ho (currently employed by Sanofi 15 Genzyme) worked on this study while employed by Geisinger. 16 17 Acronyms: 18 T1DM: type 1 diabetes mellitus; T2DM: type 2 diabetes mellitus; SU: sulfonylureas; TZD: 19 thiazolidinediones; GLP-1R: glucagon-like peptide-1 receptor; DPP4: dipeptidyl peptidase 4; SGLT2: 20 sodium-glucose cotransporter-2; MACE: Major adverse cardiovascular events; EHR: electronic health 21 record; SNVs: single nucleotide variants; GWAS: genome-wide association study 22 23 NOTE: This preprint reports new research that has not been certified by peer review and should not be used to guide clinical practice. -

Coexpression Networks Based on Natural Variation in Human Gene Expression at Baseline and Under Stress
University of Pennsylvania ScholarlyCommons Publicly Accessible Penn Dissertations Fall 2010 Coexpression Networks Based on Natural Variation in Human Gene Expression at Baseline and Under Stress Renuka Nayak University of Pennsylvania, [email protected] Follow this and additional works at: https://repository.upenn.edu/edissertations Part of the Computational Biology Commons, and the Genomics Commons Recommended Citation Nayak, Renuka, "Coexpression Networks Based on Natural Variation in Human Gene Expression at Baseline and Under Stress" (2010). Publicly Accessible Penn Dissertations. 1559. https://repository.upenn.edu/edissertations/1559 This paper is posted at ScholarlyCommons. https://repository.upenn.edu/edissertations/1559 For more information, please contact [email protected]. Coexpression Networks Based on Natural Variation in Human Gene Expression at Baseline and Under Stress Abstract Genes interact in networks to orchestrate cellular processes. Here, we used coexpression networks based on natural variation in gene expression to study the functions and interactions of human genes. We asked how these networks change in response to stress. First, we studied human coexpression networks at baseline. We constructed networks by identifying correlations in expression levels of 8.9 million gene pairs in immortalized B cells from 295 individuals comprising three independent samples. The resulting networks allowed us to infer interactions between biological processes. We used the network to predict the functions of poorly-characterized human genes, and provided some experimental support. Examining genes implicated in disease, we found that IFIH1, a diabetes susceptibility gene, interacts with YES1, which affects glucose transport. Genes predisposing to the same diseases are clustered non-randomly in the network, suggesting that the network may be used to identify candidate genes that influence disease susceptibility.