The RNA-Binding Protein Hud Regulates Alternative Splicing and Alternative Polyadenylation in the Mouse Neocortex
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mutation of the AAUAAA Polyadenylation Signal Depresses in Vitro Splicing of Proximal but Not Distal Introns
Downloaded from genesdev.cshlp.org on September 24, 2021 - Published by Cold Spring Harbor Laboratory Press Mutation of the AAUAAA polyadenylation signal depresses in vitro splicing of proximal but not distal introns Maho Niwa and Susan M. Berget Marrs McClean Department of Biochemistry, Baylor College of Medicine, Houston, Texas 77030 USA To investigate the relationship between splicing and polyadenylation during the production of vertebrate mRNAs, we examined the effect of mutation of a poly(A) site on splicing of upstream introns. Mutation of the AAUAAA polyadenylation consensus sequence inhibited in vitro splicing of an upstream intron. The magnitude of the depression depended on the magnesium concentration. Dependence of splicing on polyadenylation signals suggests the existence of interaction between polyadenylation and splicing factors. In multi-intron precursor RNAs containing duplicated splice sites, mutation of the poly(A) site inhibited removal of the last intron, but not the removal of introns farther upstream. Inhibition of removal of only the last intron suggests segmental recognition of multi-exon precursor RNAs and is consistent with previous suggestions that signals at both ends of an exon are required for effective splicing of an upstream intron. [Key Words: Polyadenylation signal; splicing; vertebrate RNAs] Received July 31, 1991; revised version accepted September 10, 1991. The majority of vertebrate mRNAs undergo both splic- Using chimeric RNAs competent for both in vitro ing and polyadenylation during maturation. The rela- splicing and polyadenylation, we recently reported that tionship between the two processing steps has been un- in vitro polyadenylation is stimulated when an intron is clear. Polyadenylation has been reported to occur shortly placed upstream of a poly(A) site (Niwa et al. -

A Protein Required for RNA Processing and Splicing in Neurospora
Proc. Nati. Acad. Sci. USA Vol. 89, pp. 1676-1680, March 1992 Genetics A protein required for RNA processing and splicing in Neurospora mitochondria is related to gene products involved in cell cycle protein phosphatase functions BEATRICE TURCQ*, KATHERINE F. DOBINSONt, NOBUFUSA SERIZAWAt, AND ALAN M. LAMBOWITZ§ Departments of Molecular Genetics and Biochemistry, and the Biotechnology Center, The Ohio State University, 484 West Twelfth Avenue, Columbus, OH 43210 Communicated by Thomas R. Cech, November 25, 1991 ABSTRACT The Neurospora crassa cyt4 mutants have specifically affect the splicing of group I introns and have pleiotropic defects in mitochondrial RNA splicing, 5' and 3' end relatively little effect on other mitochondrial RNA processing processing, and RNA turnover. Here, we show that the cyt-4 reactions (1, 4, 7). gene encodes a 120-kDa protein with significant similarity to By contrast, the cyt4 mutants have a complex phenotype the SSD1/SRK1 protein of Saceharomyces cerevisiae and the with defects in a number of different aspects of mitochondrial DIS3 protein of Sclhizosaccharomyces pombe, which have been RNA metabolism in addition to RNA splicing (9, 10). The five implicated in protein phosphatase functions that regulate cell cyt4 mutants are cold sensitive; they grow slowly at 250C and cycle and mitotic chromosome segregation. The CYT-4 protein more rapidly at 370C. Initial studies focusing on the mitochon- is present in mitochondria and is truncated or deficient in two drial large rRNA showed that the cyt4 mutants are defective cyt4 mutants. Assuming that the CYT-4 protein functions in a in both splicing and 3' end synthesis and that the splicing defect manner similar to the SSD1/SRK1 and DIS3 proteins, we infer is more pronounced at lower temperatures (9). -

Protein Interaction Network of Alternatively Spliced Isoforms from Brain Links Genetic Risk Factors for Autism
ARTICLE Received 24 Aug 2013 | Accepted 14 Mar 2014 | Published 11 Apr 2014 DOI: 10.1038/ncomms4650 OPEN Protein interaction network of alternatively spliced isoforms from brain links genetic risk factors for autism Roser Corominas1,*, Xinping Yang2,3,*, Guan Ning Lin1,*, Shuli Kang1,*, Yun Shen2,3, Lila Ghamsari2,3,w, Martin Broly2,3, Maria Rodriguez2,3, Stanley Tam2,3, Shelly A. Trigg2,3,w, Changyu Fan2,3, Song Yi2,3, Murat Tasan4, Irma Lemmens5, Xingyan Kuang6, Nan Zhao6, Dheeraj Malhotra7, Jacob J. Michaelson7,w, Vladimir Vacic8, Michael A. Calderwood2,3, Frederick P. Roth2,3,4, Jan Tavernier5, Steve Horvath9, Kourosh Salehi-Ashtiani2,3,w, Dmitry Korkin6, Jonathan Sebat7, David E. Hill2,3, Tong Hao2,3, Marc Vidal2,3 & Lilia M. Iakoucheva1 Increased risk for autism spectrum disorders (ASD) is attributed to hundreds of genetic loci. The convergence of ASD variants have been investigated using various approaches, including protein interactions extracted from the published literature. However, these datasets are frequently incomplete, carry biases and are limited to interactions of a single splicing isoform, which may not be expressed in the disease-relevant tissue. Here we introduce a new interactome mapping approach by experimentally identifying interactions between brain-expressed alternatively spliced variants of ASD risk factors. The Autism Spliceform Interaction Network reveals that almost half of the detected interactions and about 30% of the newly identified interacting partners represent contribution from splicing variants, emphasizing the importance of isoform networks. Isoform interactions greatly contribute to establishing direct physical connections between proteins from the de novo autism CNVs. Our findings demonstrate the critical role of spliceform networks for translating genetic knowledge into a better understanding of human diseases. -

Differential Analysis of Gene Expression and Alternative Splicing
i bioRxiv preprint doi: https://doi.org/10.1101/2020.08.23.234237; this version posted August 24, 2020. The copyright holder for this preprint i (which was not certified by peer review)“main” is the author/funder, — 2020/8/23 who has — granted 9:38 — bioRxiv page a1 license — #1 to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. i i EmpiReS: Differential Analysis of Gene Expression and Alternative Splicing Gergely Csaba∗, Evi Berchtold*,y, Armin Hadziahmetovic, Markus Gruber, Constantin Ammar and Ralf Zimmer Department of Informatics, Ludwig-Maximilians-Universitat¨ Munchen,¨ 80333, Munich, Germany ABSTRACT to translation. Different transcripts of the same gene are often expressed at the same time, possibly at different abundance, While absolute quantification is challenging in high- yielding a defined mixture of isoforms. Changes to this throughput measurements, changes of features composition are further referred to as differential alternative between conditions can often be determined with splicing (DAS). Various diseases can be linked to DAS high precision. Therefore, analysis of fold changes events (3). Most hallmarks of cancer like tumor progression is the standard method, but often, a doubly and immune escape (4, 5) can be attributed to changes in differential analysis of changes of changes is the transcript composition (6). DAS plays a prominent role required. Differential alternative splicing is an in various types of muscular dystrophy (MD) (7), splicing example of a doubly differential analysis, i.e. fold intervention by targeted exon removal is a therapeutic option changes between conditions for different isoforms in Duchenne MD (8). -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Transcription to RNA from Gene to Phenotype
11/8/11 Transcription to RNA From Gene to Phenotype DNA Gene 2 • The central dogma: molecule – DNA->RNA->protein Gene 1 – One Gene, One Enzyme Gene 3 • Beadle & Tatum expts. • Transcription DNA strand 3! 5! – Initiation (template) A C C A A A C C G A G T – Elongation – Termination TRANSCRIPTION U G G U U U G G C U C A • mRNA processing mRNA 5! 3! – Introns and exons Codon TRANSLATION • Other types of RNA 11/9/2011 Protein Trp Phe Gly Ser Amino acid In Prokaryotes transcription and translation occur simultaneously In Eukaryotes Nuclear – Transcription and envelope translation occur in separate compartments TRANSCRIPTION DNA DNA TRANSCRIPTION of the cell Pre-mRNA RNA PROCESSING mRNA Ribosome – RNA transcripts are mRNA TRANSLATION modified before becoming true mRNA Ribosome Polypeptide TRANSLATION Polypeptide Figure 17.3a Figure 17.3b One Gene -> One Enzyme “One Gene -> One Enzyme” EXPERIMENT Class I Class II Class III Wild type Mutants Mutants Mutants Minimal Normal bread-mold cells can medium synthesize arginine from precursors (MM) (control) in the minimal medium MM + Ornithine Mutant 2 could MM + grow if either Citrulline Precursor Ornithine Citrulline Arginine citruline or MM + arginine was Specific enzymes (arrows) Arginine is an essential Arginine amino acid, required for (control) supplied. catalyze each step growth Therefore it must lack the enzyme to make Citruline Precursor Ornithine Citrulline Arginine 1 11/8/11 From Gene to Phenotype RNA Polymerase Non-template strand of DNA DNA Gene 2 RNA nucleotides molecule Gene 1 Gene 3 T C C A A A T 3 C T U ! 3 end DNA strand 3! 5! ! G (template) A C C A A A C C G A G T T A U G G A 5 C A U C C A C TRANSCRIPTION ! A T A A G G T T U G G U U U G G C U C A mRNA 5! 3! Direction of transcription 5! Codon (“downstream) Template TRANSLATION strand of DNA Protein Trp Phe Gly Ser New RNA Amino acid Synthesis of an RNA Transcript DNA is copied to make messenger RNA Promoter Transcription unit 5! 3! 3! 5! RNA polymerase DNA This is the “non-template” Start point binds to a promoter RNA polymerase strand. -

Supplementary Materials
1 Supplementary Materials: Supplemental Figure 1. Gene expression profiles of kidneys in the Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice. (A) A heat map of microarray data show the genes that significantly changed up to 2 fold compared between Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice (N=4 mice per group; p<0.05). Data show in log2 (sample/wild-type). 2 Supplemental Figure 2. Sting signaling is essential for immuno-phenotypes of the Fcgr2b-/-lupus mice. (A-C) Flow cytometry analysis of splenocytes isolated from wild-type, Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice at the age of 6-7 months (N= 13-14 per group). Data shown in the percentage of (A) CD4+ ICOS+ cells, (B) B220+ I-Ab+ cells and (C) CD138+ cells. Data show as mean ± SEM (*p < 0.05, **p<0.01 and ***p<0.001). 3 Supplemental Figure 3. Phenotypes of Sting activated dendritic cells. (A) Representative of western blot analysis from immunoprecipitation with Sting of Fcgr2b-/- mice (N= 4). The band was shown in STING protein of activated BMDC with DMXAA at 0, 3 and 6 hr. and phosphorylation of STING at Ser357. (B) Mass spectra of phosphorylation of STING at Ser357 of activated BMDC from Fcgr2b-/- mice after stimulated with DMXAA for 3 hour and followed by immunoprecipitation with STING. (C) Sting-activated BMDC were co-cultured with LYN inhibitor PP2 and analyzed by flow cytometry, which showed the mean fluorescence intensity (MFI) of IAb expressing DC (N = 3 mice per group). 4 Supplemental Table 1. Lists of up and down of regulated proteins Accession No. -

Polymerse Activity
University of Kentucky UKnowledge University of Kentucky Doctoral Dissertations Graduate School 2005 CHARACTERIZATION OF PLANT POLYADENYLATION TRANSACTING FACTORS-FACTORS THAT MODIFY POLY(A) POLYMERSE ACTIVITY Kevin Patrick Forbes University of Kentucky Right click to open a feedback form in a new tab to let us know how this document benefits ou.y Recommended Citation Forbes, Kevin Patrick, "CHARACTERIZATION OF PLANT POLYADENYLATION TRANSACTING FACTORS- FACTORS THAT MODIFY POLY(A) POLYMERSE ACTIVITY" (2005). University of Kentucky Doctoral Dissertations. 444. https://uknowledge.uky.edu/gradschool_diss/444 This Dissertation is brought to you for free and open access by the Graduate School at UKnowledge. It has been accepted for inclusion in University of Kentucky Doctoral Dissertations by an authorized administrator of UKnowledge. For more information, please contact [email protected]. ABSTRACT OF DISSERTATION Kevin Patrick Forbes The Graduate School University of Kentucky 2004 CHARACTERIZATION OF PLANT POLYADENYLATION TRANS- ACTING FACTORS-FACTORS THAT MODIFY POLY(A) POLYMERSE ACTIVITY _________________________________________ ABSTRACT OF DISSERTATION _________________________________________ A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the College of Agriculture at the University of Kentucky By Kevin Patrick Forbes Lexington, Kentucky Director: Dr. Arthur G. Hunt, Professor of Agronomy Lexington, Kentucky 2004 Copyright ” Kevin Patrick Forbes 2004 ABSTRACT OF DISSERTATION CHARACTERIZATION OF PLANT POLYADENYLATION TRANS-ACTING FACTORS-FACTORS THAT MODIFY POLY(A) POLYMERSE ACTIVITY Plant polyadenylation factors have proven difficult to purify and characterize, owing to the presence of excessive nuclease activity in plant nuclear extracts, thereby precluding the identification of polyadenylation signal-dependent processing and polyadenylation in crude extracts. -

Supplementary Figures 1-14 and Supplementary References
SUPPORTING INFORMATION Spatial Cross-Talk Between Oxidative Stress and DNA Replication in Human Fibroblasts Marko Radulovic,1,2 Noor O Baqader,1 Kai Stoeber,3† and Jasminka Godovac-Zimmermann1* 1Division of Medicine, University College London, Center for Nephrology, Royal Free Campus, Rowland Hill Street, London, NW3 2PF, UK. 2Insitute of Oncology and Radiology, Pasterova 14, 11000 Belgrade, Serbia 3Research Department of Pathology and UCL Cancer Institute, Rockefeller Building, University College London, University Street, London WC1E 6JJ, UK †Present Address: Shionogi Europe, 33 Kingsway, Holborn, London WC2B 6UF, UK TABLE OF CONTENTS 1. Supplementary Figures 1-14 and Supplementary References. Figure S-1. Network and joint spatial razor plot for 18 enzymes of glycolysis and the pentose phosphate shunt. Figure S-2. Correlation of SILAC ratios between OXS and OAC for proteins assigned to the SAME class. Figure S-3. Overlap matrix (r = 1) for groups of CORUM complexes containing 19 proteins of the 49-set. Figure S-4. Joint spatial razor plots for the Nop56p complex and FIB-associated complex involved in ribosome biogenesis. Figure S-5. Analysis of the response of emerin nuclear envelope complexes to OXS and OAC. Figure S-6. Joint spatial razor plots for the CCT protein folding complex, ATP synthase and V-Type ATPase. Figure S-7. Joint spatial razor plots showing changes in subcellular abundance and compartmental distribution for proteins annotated by GO to nucleocytoplasmic transport (GO:0006913). Figure S-8. Joint spatial razor plots showing changes in subcellular abundance and compartmental distribution for proteins annotated to endocytosis (GO:0006897). Figure S-9. Joint spatial razor plots for 401-set proteins annotated by GO to small GTPase mediated signal transduction (GO:0007264) and/or GTPase activity (GO:0003924). -
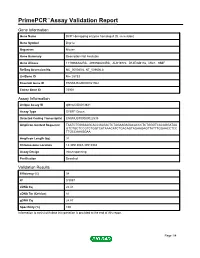
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name DCP1 decapping enzyme homolog A (S. cerevisiae) Gene Symbol Dcp1a Organism Mouse Gene Summary Description Not Available Gene Aliases 1110066A22Rik, 4930568L04Rik, AU019772, D14Ertd817e, Mitc1, SMIF RefSeq Accession No. NC_000080.6, NT_039606.8 UniGene ID Mm.28733 Ensembl Gene ID ENSMUSG00000021962 Entrez Gene ID 75901 Assay Information Unique Assay ID qMmuCID0013841 Assay Type SYBR® Green Detected Coding Transcript(s) ENSMUST00000022535 Amplicon Context Sequence TAATCTGGGAAGCACCGAGACTCTAGAAGAGACACCCTCTGGGTCACAGGATAA GTCTGCTCCGTCTGGTCATAAACATCTGACAGTAGAAGAGTTATTTGGAACCTCC TTGCCAAAGGAA Amplicon Length (bp) 91 Chromosome Location 14:30513043-30518984 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 98 R2 0.9997 cDNA Cq 22.41 cDNA Tm (Celsius) 81 gDNA Cq 24.87 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report Dcp1a, Mouse Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Mouse Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. Detected Coding Transcript(s) This is a list of the Ensembl transcript ID(s) that this assay will detect. -

Exon Amplification: a Strategy to Isolate Mammalian Genes Based on RNA Splicing (Gene Cloning/Polymerase Chain Reaction) ALAN J
Proc. Natl. Acad. Sci. USA Vol. 88, pp. 4005-4009, May 1991 Genetics Exon amplification: A strategy to isolate mammalian genes based on RNA splicing (gene cloning/polymerase chain reaction) ALAN J. BUCKLER*, DAVID D. CHANG, SHARON L. GRAw, J. DAVID BROOK, DANIEL A. HABER, PHILLIP A. SHARP, AND DAVID E. HOUSMAN Center for Cancer Research, Department of Biology, Massachusetts Institute of Technology, Cambridge, MA 02139 Contributed by Phillip A. Sharp, January 25, 1991 ABSTRACT We have developed a method, exon amplifi- (7, 8). Thus, this method may be generally applicable for the cation, for fast and efficient isolation of coding sequences from selection of exon sequences from any gene. The method is complex mammalian genomic DNA. This method is based on also both rapid and easily adapted to large scale experiments. the selection of RNA sequences, exons, which are flanked by A series of cloned genomic DNA fragments can be screened functional 5' and 3' splice sites. Fragments of cloned genomic within 1-2 weeks. The sensitivity of this method is high. DNA are inserted into an intron, which is flanked by 5' and 3' Genomic DNA segments of 20 kilobases (kb) or more can be splice sites of the human immunodeficiency virus 1 tat gene successfully screened in a single transfection by using a set contained within the plasmid pSPL1. COS-7 cells are trans- of pooled subclones. This method thus allows the rapid fected with these constructs, and the resulting RNA transcripts identification of exons in mammalian genomic DNA and are processed in vivo. Splice sites of exons contained within the should facilitate the isolation of a wide spectrum of genes of inserted genomic fragment are paired with splice sites of the significance in physiology and development. -

Mrna Turnover Philip Mitchell* and David Tollervey†
320 mRNA turnover Philip Mitchell* and David Tollervey† Nuclear RNA-binding proteins can record pre-mRNA are cotransported to the cytoplasm with the mRNP. These processing events in the structure of messenger proteins may preserve a record of the nuclear history of the ribonucleoprotein particles (mRNPs). During initial rounds of pre-mRNA in the cytoplasmic mRNP structure. This infor- translation, the mature mRNP structure is established and is mation can strongly influence the cytoplasmic fate of the monitored by mRNA surveillance systems. Competition for the mRNA and is used by mRNA surveillance systems that act cap structure links translation and subsequent mRNA as a checkpoint of mRNP integrity, particularly in the identi- degradation, which may also involve multiple deadenylases. fication of premature translation termination codons (PTCs). Addresses Cotransport of nuclear mRNA-binding proteins with mRNA Wellcome Trust Centre for Cell Biology, ICMB, University of Edinburgh, from the nucleus to the cytoplasm (nucleocytoplasmic shut- Kings’ Buildings, Edinburgh EH9 3JR, UK tling) was first observed for the heterogeneous nuclear *e-mail: [email protected] ribonucleoprotein (hnRNP) proteins. Some hnRNP proteins †e-mail: [email protected] are stripped from the mRNA at export [1], but hnRNP A1, Current Opinion in Cell Biology 2001, 13:320–325 A2, E, I and K are all exported (see [2]). Although roles for 0955-0674/01/$ — see front matter these hnRNP proteins in transport and translation have been © 2001 Elsevier Science Ltd. All rights reserved. reported [3•,4•], their affects on mRNA stability have been little studied. More is known about hnRNP D/AUF1 and Abbreviations AREs AU-rich sequence elements another nuclear RNA-binding protein, HuR, which act CBC cap-binding complex antagonistically to modulate the stability of a range of DAN deadenylating nuclease mRNAs containing AU-rich sequence elements (AREs) DSEs downstream sequence elements (reviewed in [2]).