Single-Molecule Study of Β-Catenin Translocation And
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplemental Material
Supplemental Table B ARGs in alphabetical order Symbol Title 3 months 6 months 9 months 12 months 23 months ANOVA Direction Category 38597 septin 2 1557 ± 44 1555 ± 44 1579 ± 56 1655 ± 26 1691 ± 31 0.05219 up Intermediate 0610031j06rik kidney predominant protein NCU-G1 491 ± 6 504 ± 14 503 ± 11 527 ± 13 534 ± 12 0.04747 up Early Adult 1G5 vesicle-associated calmodulin-binding protein 662 ± 23 675 ± 17 629 ± 16 617 ± 20 583 ± 26 0.03129 down Intermediate A2m alpha-2-macroglobulin 262 ± 7 272 ± 8 244 ± 6 290 ± 7 353 ± 16 0.00000 up Midlife Aadat aminoadipate aminotransferase (synonym Kat2) 180 ± 5 201 ± 12 223 ± 7 244 ± 14 275 ± 7 0.00000 up Early Adult Abca2 ATP-binding cassette, sub-family A (ABC1), member 2 958 ± 28 1052 ± 58 1086 ± 36 1071 ± 44 1141 ± 41 0.05371 up Early Adult Abcb1a ATP-binding cassette, sub-family B (MDR/TAP), member 1A 136 ± 8 147 ± 6 147 ± 13 155 ± 9 185 ± 13 0.01272 up Midlife Acadl acetyl-Coenzyme A dehydrogenase, long-chain 423 ± 7 456 ± 11 478 ± 14 486 ± 13 512 ± 11 0.00003 up Early Adult Acadvl acyl-Coenzyme A dehydrogenase, very long chain 426 ± 14 414 ± 10 404 ± 13 411 ± 15 461 ± 10 0.01017 up Late Accn1 amiloride-sensitive cation channel 1, neuronal (degenerin) 242 ± 10 250 ± 9 237 ± 11 247 ± 14 212 ± 8 0.04972 down Late Actb actin, beta 12965 ± 310 13382 ± 170 13145 ± 273 13739 ± 303 14187 ± 269 0.01195 up Midlife Acvrinp1 activin receptor interacting protein 1 304 ± 18 285 ± 21 274 ± 13 297 ± 21 341 ± 14 0.03610 up Late Adk adenosine kinase 1828 ± 43 1920 ± 38 1922 ± 22 2048 ± 30 1949 ± 44 0.00797 up Early -

Autoantibodies and Anti-Microbial Antibodies
bioRxiv preprint doi: https://doi.org/10.1101/403519; this version posted August 29, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Autoantibodies and anti-microbial antibodies: Homology of the protein sequences of human autoantigens and the microbes with implication of microbial etiology in autoimmune diseases Peilin Zhang, MD., Ph.D. PZM Diagnostics, LLC Charleston, WV 25301 Correspondence: Peilin Zhang, MD., Ph.D. PZM Diagnostics, LLC. 500 Donnally St., Suite 303 Charleston, WV 25301 Email: [email protected] Tel: 304 444 7505 1 bioRxiv preprint doi: https://doi.org/10.1101/403519; this version posted August 29, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Abstract Autoimmune disease is a group of diverse clinical syndromes with defining autoantibodies within the circulation. The pathogenesis of autoantibodies in autoimmune disease is poorly understood. In this study, human autoantigens in all known autoimmune diseases were examined for the amino acid sequences in comparison to the microbial proteins including bacterial and fungal proteins by searching Genbank protein databases. Homologies between the human autoantigens and the microbial proteins were ranked high, medium, and low based on the default search parameters at the NCBI protein databases. Totally 64 human protein autoantigens important for a variety of autoimmune diseases were examined, and 26 autoantigens were ranked high, 19 ranked medium to bacterial proteins (69%) and 27 ranked high and 16 ranked medium to fungal proteins (66%) in their respective amino acid sequence homologies. -

Mutations in Nucleoporin NUP88 Cause Lethal Neuromuscular Disorder
bioRxiv preprint doi: https://doi.org/10.1101/347179; this version posted August 14, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Mutations in nucleoporin NUP88 cause lethal neuromuscular disorder Edith Bonnin1,2,#, Pauline Cabochette2,#, Alessandro Filosa3, Ramona Jühlen1, Shoko Komatsuzaki4,§, Mohammed Hezwani1, Achim Dickmanns5, Valérie Martinelli1, Marjorie Vermeersch6, Lynn Supply7, Nuno Martins1, Laurence Pirenne1, Gianina Ravenscroft8, Marcus Lombard8, Sarah Port9,§§, Christiane Spillner9, Sandra Janssens10, Ellen Roets11, Jo Van Dorpe7, Martin Lammens12, Ralph H. Kehlenbach9, Ralf Ficner5, Nigel Laing8, Katrin Hoffmann4, Benoit Vanhollebeke2, *, and Birthe Fahrenkrog1,* 1Institute for Molecular Biology and Medicine, Laboratory Biology of the Cell Nucleus, Université Libre de Bruxelles (ULB), 6041 Gosselies, Belgium; 2Laboratory of Neurovascular Signaling, Department of Molecular Biology, ULB Neuroscience Institute, Université Libre de Bruxelles (ULB), 6041 Gosselies, Belgium; 3Max Delbrück Center for Molecular Medicine, 13092 Berlin, Germany; 4Institute of Human Genetics, Martin-Luther-University Halle-Wittenberg, 06112 Halle, Germany; 5Department of Molecular Structural Biology, Institute for Microbiology and Genetics, GZMB, Georg-August-University Göttingen, 37077 Göttingen, Germany; 6Center for Microscopy and Molecular Imaging, -

The Genetic Program of Pancreatic Beta-Cell Replication in Vivo
Page 1 of 65 Diabetes The genetic program of pancreatic beta-cell replication in vivo Agnes Klochendler1, Inbal Caspi2, Noa Corem1, Maya Moran3, Oriel Friedlich1, Sharona Elgavish4, Yuval Nevo4, Aharon Helman1, Benjamin Glaser5, Amir Eden3, Shalev Itzkovitz2, Yuval Dor1,* 1Department of Developmental Biology and Cancer Research, The Institute for Medical Research Israel-Canada, The Hebrew University-Hadassah Medical School, Jerusalem 91120, Israel 2Department of Molecular Cell Biology, Weizmann Institute of Science, Rehovot, Israel. 3Department of Cell and Developmental Biology, The Silberman Institute of Life Sciences, The Hebrew University of Jerusalem, Jerusalem 91904, Israel 4Info-CORE, Bioinformatics Unit of the I-CORE Computation Center, The Hebrew University and Hadassah, The Institute for Medical Research Israel- Canada, The Hebrew University-Hadassah Medical School, Jerusalem 91120, Israel 5Endocrinology and Metabolism Service, Department of Internal Medicine, Hadassah-Hebrew University Medical Center, Jerusalem 91120, Israel *Correspondence: [email protected] Running title: The genetic program of pancreatic β-cell replication 1 Diabetes Publish Ahead of Print, published online March 18, 2016 Diabetes Page 2 of 65 Abstract The molecular program underlying infrequent replication of pancreatic beta- cells remains largely inaccessible. Using transgenic mice expressing GFP in cycling cells we sorted live, replicating beta-cells and determined their transcriptome. Replicating beta-cells upregulate hundreds of proliferation- related genes, along with many novel putative cell cycle components. Strikingly, genes involved in beta-cell functions, namely glucose sensing and insulin secretion were repressed. Further studies using single molecule RNA in situ hybridization revealed that in fact, replicating beta-cells double the amount of RNA for most genes, but this upregulation excludes genes involved in beta-cell function. -

Mapping Transmembrane Binding Partners for E-Cadherin Ectodomains
SUPPLEMENTARY INFORMATION TITLE: Mapping transmembrane binding partners for E-cadherin ectodomains. AUTHORS: Omer Shafraz 1, Bin Xie 2, Soichiro Yamada 1, Sanjeevi Sivasankar 1, 2, * AFFILIATION: 1 Department of Biomedical Engineering, 2 Biophysics Graduate Group, University of California, Davis, CA 95616. *CORRESPONDING AUTHOR: Sanjeevi Sivasankar, Tel: (530)-754-0840, Email: [email protected] Figure S1: Western blots a. EC-BioID, WT and Ecad-KO cell lysates stained for Ecad and tubulin. b. HRP-streptavidin staining of biotinylated proteins eluted from streptavidin coated magnetic beads incubated with cell lysates of EC-BioID with (+) and without (-) exogenous biotin. c. C-BioID, WT and Ecad-KO cell lysates stained for Ecad and tubulin. d. HRP-streptavidin staining of biotinylated proteins eluted from streptavidin coated magnetic beads incubated with cell lysates of C-BioID with (+) and without (-) exogenous biotin. (+) Biotin (-) Biotin Sample 1 Sample 2 Sample 3 Sample 4 Sample 1 Sample 2 Sample 3 Sample 4 Percent Percent Percent Percent Percent Percent Percent Percent Gene ID Coverage Coverage Coverage Coverage Coverage Coverage Coverage Coverage CDH1 29.6 31.4 41.1 36.5 10.8 6.7 28.8 29.1 DSG2 26 14.6 45 37 0.8 1.9 1.6 18.7 CXADR 30.2 26.2 32.7 27.1 0.0 0.0 0.0 6.9 EFNB1 24.3 30.6 24 30.3 0.0 0.0 0.0 0.0 ITGA2 16.5 22.2 30.1 33.4 1.1 1.1 5.2 7.2 CDH3 21.8 9.7 20.6 25.3 1.3 1.3 0.0 0.0 ITGB1 11.8 16.7 23.9 20.3 0.0 2.9 8.5 5.8 DSC3 9.7 7.5 11.5 13.3 0.0 0.0 2.6 0.0 EPHA2 23.2 31.6 31.6 30.5 0.8 0.0 0.0 5.7 ITGB4 21.8 27.8 33.1 30.7 0.0 1.2 3.9 4.4 ITGB3 23.5 22.2 26.8 24.7 0.0 0.0 5.2 9.1 CDH6 22.8 18.1 28.6 24.3 0.0 0.0 0.0 9.1 CDH17 8.8 12.4 20.7 18.4 0.0 0.0 0.0 0.0 ITGB6 12.7 10.4 14 17.1 0.0 0.0 0.0 1.7 EPHB4 11.4 8.1 14.2 16.3 0.0 0.0 0.0 0.0 ITGB8 5 10 15 17.6 0.0 0.0 0.0 0.0 ITGB5 6.2 9.5 15.2 13.8 0.0 0.0 0.0 0.0 EPHB2 8.5 4.8 9.8 12.1 0.0 0.0 0.0 0.0 CDH24 5.9 7.2 8.3 9 0.0 0.0 0.0 0.0 Table S1: EC-BioID transmembrane protein hits. -

New Activities of the Nuclear Pore Complexes
cells Editorial New Activities of the Nuclear Pore Complexes Richard W. Wong 1,2,3 1 WPI-Nano Life Science Institute, Kanazawa University, Kanazawa 920-1192, Japan; [email protected] 2 Graduate School of Frontier Science Initiative, Kanazawa University, Kanazawa 920-1192, Japan 3 Cell-Bionomics Research Unit, Institute for Frontier Science Initiative, Kanazawa University, Kanazawa 920-1192, Japan Abstract: Nuclear pore complexes (NPCs) at the surface of nuclear membranes play a critical role in regulating the transport of both small molecules and macromolecules between the cell nucleus and cytoplasm via their multilayered spiderweb-like central channel. During mitosis, nuclear envelope breakdown leads to the rapid disintegration of NPCs, allowing some NPC proteins to play crucial roles in the kinetochore structure, spindle bipolarity, and centrosome homeostasis. The aberrant functioning of nucleoporins (Nups) and NPCs has been associated with autoimmune diseases, viral infections, neurological diseases, cardiomyopathies, and cancers, especially leukemia. This Special Issue highlights several new contributions to the understanding of NPC proteostasis. Keywords: nuclear pore complex; nanomedicine; liquid–liquid phase separation; biomacromolecule; HS-AFM; nucleoporin; nanoimaging The nuclear pore complex (NPC) [1–3] is a nanoscale gatekeeper with a central, se- lective, cobweb-like barrier composed mainly of nucleoporins (Nups), which comprise intrinsically disordered (non-structured) regions (IDRs) with phenylalanine–glycine (FG) motifs (FG-Nups) [4–8]. A fully assembled NPC in vertebrates contains multiple copies of approximately 30 different Nups and has an estimated molecular mass of 120 MDa [9]. Citation: Wong, R.W. New Activities Despite our knowledge of the NPC structure, the molecular mechanisms underlying of the Nuclear Pore Complexes. -

Immunity in Primary Biliary Cirrhosis
Genes and Immunity (2005) 6, 543–556 & 2005 Nature Publishing Group All rights reserved 1466-4879/05 $30.00 www.nature.com/gene REVIEW Genes and (auto)immunity in primary biliary cirrhosis C Selmi1,2, P Invernizzi1, M Zuin1, M Podda1, MF Seldin2,3 and ME Gershwin2 1Division of Internal Medicine, San Paolo School of Medicine, University of Milan, Milan, Italy; 2Division of Rheumatology, Allergy and Clinical Immunology, University of California at Davis, Davis, CA 95616, USA; 3Biochemistry and Rowe Program in Human Genetics, University of California at Davis, School of Medicine, Davis, CA, USA Primary biliary cirrhosis (PBC) is a chronic autoimmune cholestatic liver disease most commonly encountered in postmenopausal women; it is characterized by high-titer serum autoantibodies to mitochondrial antigens, elevated serum IgM, progressive destruction of intrahepatic bile ducts, and ultimately liver cirrhosis and failure. The cytopathic mechanisms leading to the selective destruction of intrahepatic cholangiocytes are still largely unknown. The current theory on the pathogenesis of PBC indicated that environmental factors might trigger autoimmunity in genetically susceptible individuals. In fact, genetic predisposition is critical to disease onset and progression, yet peculiar among autoimmune diseases, as indicated by the lack of a strong association with major histocompatibility complex haplotypes. Further, the recently reported concordance rate among monozygotic twins strenghtens the importance of genetic factors, while also indicating that additional factors, possibly infectious agents or xenobiotics, intervene to trigger the disease. In this review, the available data regarding the genetic factors associated with PBC susceptibility and progression, as well as the available evidence regarding the immunomediated pathogenesis of PBC, will be critically illustrated and discussed. -

The Stoichiometry of the Nucleoporin 62 Subcomplex of the Nuclear Pore in Solution
The stoichiometry of the nucleoporin 62 subcomplex of the nuclear pore in solution The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation Ulrich, A., J. R. Partridge, and T. U. Schwartz. “The Stoichiometry of the Nucleoporin 62 Subcomplex of the Nuclear Pore in Solution.” Molecular Biology of the Cell 25, no. 9 (May 1, 2014): 1484–1492. As Published http://dx.doi.org/10.1091/mbc.E13-12-0745 Publisher American Society for Cell Biology Version Final published version Citable link http://hdl.handle.net/1721.1/87562 Terms of Use Article is made available in accordance with the publisher's policy and may be subject to US copyright law. Please refer to the publisher's site for terms of use. M BoC | ARTICLE The stoichiometry of the nucleoporin 62 subcomplex of the nuclear pore in solution Alexander Ulricha,b, James R. Partridgea, and Thomas U. Schwartza aDepartment of Biology, Massachusetts Institute of Technology, Cambridge, MA 02139; bInstitut für Chemie und Biochemie, AG Strukturbiochemie, Freie Universität Berlin, 14195 Berlin, Germany ABSTRACT The nuclear pore complex (NPC) regulates transport between the nucleus and Monitoring Editor cytoplasm. Soluble cargo-protein complexes navigate through the pore by binding to pheny- Martin Hetzer lalanine-glycine (FG)-repeat proteins attached to the channel walls. The Nup62 complex con- Salk Institute for Biological Studies tains the FG-repeat proteins Nup62, Nup54, and Nup58 and is located in the center of the NPC. The three proteins bind each other via conserved coiled-coil segments. To determine Received: Dec 19, 2013 the stoichiometry of the Nup62 complex, we undertook an in vitro study using gel filtration Revised: Feb 12, 2014 and analytical ultracentrifugation. -

Host-HIV-1 Interactome: a Quest for Novel Therapeutic Intervention
cells Review Host-HIV-1 Interactome: A Quest for Novel Therapeutic Intervention Ekta Shukla and Radha Chauhan * National Center for Cell Science, S.P Pune University, Pune-411007, Maharashtra, India; [email protected] * Correspondence: [email protected]; Tel.: +91-20-25708251 Received: 16 August 2019; Accepted: 9 September 2019; Published: 27 September 2019 Abstract: The complex nature and structure of the human immunodeficiency virus has rendered the cure for HIV infections elusive. The advances in antiretroviral treatment regimes and the development of highly advanced anti-retroviral therapy, which primarily targets the HIV enzymes, have dramatically changed the face of the HIV epidemic worldwide. Despite this remarkable progress, patients treated with these drugs often witness inadequate efficacy, compound toxicity and non-HIV complications. Considering the limited inventory of druggable HIV proteins and their susceptibility to develop drug resistance, recent attempts are focussed on targeting HIV-host interactomes that are essential for viral reproduction. Noticeably, unlike other viruses, HIV subverts the host nuclear pore complex to enter into and exit through the nucleus. Emerging evidence suggests a crucial role of interactions between HIV-1 proteins and host nucleoporins that underlie the import of the pre-integration complex into the nucleus and export of viral RNAs into the cytoplasm during viral replication. Nevertheless, the interaction of HIV-1 with nucleoporins has been poorly described and the role of nucleoporins during nucleocytoplasmic transport of HIV-1 still remains unclear. In this review, we highlight the advances and challenges in developing a more effective antiviral arsenal by exploring critical host-HIV interactions with a special focus on nuclear pore complex (NPC) and nucleoporins. -
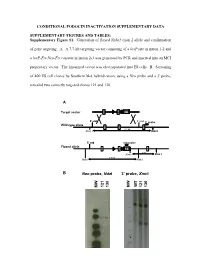
Supplementary Figure S1. Generation of Floxed Nphs2 Exon 2 Allele and Confirmation
CONDITIONAL PODOCIN INACTIVATION SUPPLEMENTARY DATA SUPPLEMENTARY FIGURES AND TABLES: Supplementary Figure S1. Generation of floxed Nphs2 exon 2 allele and confirmation of gene targeting. A. A 7.7-kb targeting vector consisting of a loxP site in intron 1-2 and a loxP-Frt-Neo-Frt cassette in intron 2-3 was generated by PCR and inserted into an MCI proprietary vector. The linearized vector was electroporated into ES cells. B. Screening of 400 ES cell clones by Southern blot hybridization, using a Neo probe and a 3' probe, revealed two correctly targeted clones 121 and 130. A Target vector 1 2 NEO 5’ end 3’ end 3’ probe Wild-type allele 1 2 3 9.2 kb Xmn I Xmn I 5’ end neo probe Floxed allele 1 2 NEO 3 5 kb Xmn I Xmn I 7.8 kb Nde I Nde I B Neo probe, NdeI 3’ probe, XmnI WT 121 130 121 130 MW MW - 1 - Supplementary Figure S2. Generation of Nphs2lox2/-,Cre+ mice and excision of exon 2 upon Cre recombinase induction. A. Triallelic Nphs2lox2/-,Cre+ mice were obtained by mating phenotypically normal Nphs2lox2/lox2 mice with Nphs2+/-,Cre+ mice. Mendelian inheritance of these alleles was observed. B. Genotypes were verified by multiplex PCR of tail genomic DNA. C. Cre recombinase activity was induced upon tamoxifen administration, leading to excision of the floxed exon 2 of the Nphs2 gene. D. Cre recombinase activity in the kidney was verified by PCR using a set of forward and reverse primers designed around exon 2 and demonstrating a 692-bp product (before Cre) and a 316-bp product (after Cre) using genomic DNA extracted from the renal cortex. -

Noncovalent SUMO-Interaction Motifs in HIV Integrase Play Important
Zheng et al. Virology Journal (2019) 16:42 https://doi.org/10.1186/s12985-019-1134-8 RESEARCH Open Access Noncovalent SUMO-interaction motifs in HIV integrase play important roles in SUMOylation, cofactor binding, and virus replication Yingfeng Zheng1, Kallesh Danappa Jayappa1, Zhujun Ao1,2, Xiangguo Qiu2, Ruey-Chyi Su2 and Xiaojian Yao1,2* Abstract Background: HIV integrase (IN) and its cellular cofactors, including lens-epithelium-derived growth factor (LEDGF/ p75), Ku70, p300, and Rad52, are subject to small ubiquitin-like modifier (SUMO) modification. In addition to covalent SUMOylation, SUMO paralogs can also noncovalently bind proteins through SUMO-interacting motifs (SIMs). However, little is known about whether HIV IN contains SIMs and the roles of these motifs. Results: We searched for the amino acid sequence of HIV IN and investigated three putative SIMs of IN: SIM1 72VILV75, SIM2 200IVDI203 and SIM3 257IKVV260. Our mutational analysis showed that 200IVDI203 and 257IKVV260 are two bona fide SIMs that mediate IN-SUMO noncovalent interactions. Additionally, a cell-based SUMOylation assay revealed that IN SIMs negatively regulate the SUMOylation of IN, as well as the interaction between IN and SUMO E2 conjugation enzyme Ubc9. Conversely, IN SIMs are required for its interactions with LEDGF/p75 but not with Ku70. Furthermore, our study reveals that SIM2 and SIM3 are required for the nuclear localization of IN. Finally, we investigated the impact of IN SIM2 and SIM3 on HIV single cycle replication in CD4+ C8166 T cells, and the results showed that viruses carrying IN SIM mutants are replication defective at the steps of the early viral life cycle, including reverse transcription, nuclear import and integration. -

A Meta-Analysis of the Effects of High-LET Ionizing Radiations in Human Gene Expression
Supplementary Materials A Meta-Analysis of the Effects of High-LET Ionizing Radiations in Human Gene Expression Table S1. Statistically significant DEGs (Adj. p-value < 0.01) derived from meta-analysis for samples irradiated with high doses of HZE particles, collected 6-24 h post-IR not common with any other meta- analysis group. This meta-analysis group consists of 3 DEG lists obtained from DGEA, using a total of 11 control and 11 irradiated samples [Data Series: E-MTAB-5761 and E-MTAB-5754]. Ensembl ID Gene Symbol Gene Description Up-Regulated Genes ↑ (2425) ENSG00000000938 FGR FGR proto-oncogene, Src family tyrosine kinase ENSG00000001036 FUCA2 alpha-L-fucosidase 2 ENSG00000001084 GCLC glutamate-cysteine ligase catalytic subunit ENSG00000001631 KRIT1 KRIT1 ankyrin repeat containing ENSG00000002079 MYH16 myosin heavy chain 16 pseudogene ENSG00000002587 HS3ST1 heparan sulfate-glucosamine 3-sulfotransferase 1 ENSG00000003056 M6PR mannose-6-phosphate receptor, cation dependent ENSG00000004059 ARF5 ADP ribosylation factor 5 ENSG00000004777 ARHGAP33 Rho GTPase activating protein 33 ENSG00000004799 PDK4 pyruvate dehydrogenase kinase 4 ENSG00000004848 ARX aristaless related homeobox ENSG00000005022 SLC25A5 solute carrier family 25 member 5 ENSG00000005108 THSD7A thrombospondin type 1 domain containing 7A ENSG00000005194 CIAPIN1 cytokine induced apoptosis inhibitor 1 ENSG00000005381 MPO myeloperoxidase ENSG00000005486 RHBDD2 rhomboid domain containing 2 ENSG00000005884 ITGA3 integrin subunit alpha 3 ENSG00000006016 CRLF1 cytokine receptor like