New Insights Into the Systematics of Fish Genus Gaidropsarus (Gadiformes, Gadidae): Flagging Synonymies and Hidden Diversity
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Histological Aspects of the Early Development of the Digestive System of Burbot Lota Lota L
ISSN 0015-5497, e-ISSN 1734-9168 Folia Biologica (Kraków), vol. 64 (2016), No 1 Ó Institute of Systematics and Evolution of Animals, PAS, Kraków, 2015 doi:10.3409/fb64_1.11 Histological Aspects of the Early Development of the Digestive System of Burbot Lota lota L. (Lotidae, Gadiformes)* Gra¿yna FURGA£A-SELEZNIOW, Ma³gorzata JANKUN, Roman KUJAWA, Joanna NOWOSAD, Maria BI£AS, Dariusz KUCHARCZYK, and Andrzej SKRZYPCZAK Accepted December 04, 2015 Published January 29, 2016 FURGA£A-SELEZNIOW G., JANKUN M., KUJAWA R., NOWOSAD J., BI£AS M., KUCHARCZYK D., SKRZYPCZAK A. 2016. Histological aspects of the early development of the digestive system of burbot Lota lota L. (Lotidae, Gadiformes). Folia Biologica (Kraków) 64: 11-21. The ontogeny of the digestive tract was studied histologically in burbot, Lota lota L., from hatching to 42 days post-hatch (dph). At hatching, the digestive tract consisted of a straight tube with discernible digestive accessory glands (the liver and the pancreas) dorsally attached to the yolk sac. Most of the yolk sac reserves were consumed during the first 12 days and were completely depleted by 17 dph. The first PAS-positive goblet cells appeared at 6 dph, dispersed within the epithelium of the oesophagus and increasing substantially in number and distribution as development progressed. At 12 dph, the first vacuoles (neutral lipids) appeared in the intestine, indicating the functional absorption of nutrients from food. Differentiation of gastric glands was first noticed at 17 dph and was extensive by 27 dph. L. lota larvae have a morphologically complete digestive tract by 32 dph. -
![Secondary Sexual Characteristics in Codfishes ([[Gadidae]]) in Relation to Sound Production, Habitat Use, and Social Behaviour](https://docslib.b-cdn.net/cover/5574/secondary-sexual-characteristics-in-codfishes-gadidae-in-relation-to-sound-production-habitat-use-and-social-behaviour-325574.webp)
Secondary Sexual Characteristics in Codfishes ([[Gadidae]]) in Relation to Sound Production, Habitat Use, and Social Behaviour
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by International Institute for Applied Systems Analysis (IIASA) Secondary sexual characteristics in codfishes ([[Gadidae]]) in relation to sound production, habitat use, and social behaviour Skjaeraasen, J.E., Meager, J.J. and Heino, M. IIASA Interim Report 2012 Skjaeraasen, J.E., Meager, J.J. and Heino, M. (2012) Secondary sexual characteristics in codfishes ([[Gadidae]]) in relation to sound production, habitat use, and social behaviour. IIASA Interim Report. IR-12-071 Copyright © 2012 by the author(s). http://pure.iiasa.ac.at/10208/ Interim Report on work of the International Institute for Applied Systems Analysis receive only limited review. Views or opinions expressed herein do not necessarily represent those of the Institute, its National Member Organizations, or other organizations supporting the work. All rights reserved. Permission to make digital or hard copies of all or part of this work for personal or classroom use is granted without fee provided that copies are not made or distributed for profit or commercial advantage. All copies must bear this notice and the full citation on the first page. For other purposes, to republish, to post on servers or to redistribute to lists, permission must be sought by contacting [email protected] International Institute for Tel: +43 2236 807 342 Applied Systems Analysis Fax: +43 2236 71313 Schlossplatz 1 E-mail: [email protected] A-2361 Laxenburg, Austria Web: www.iiasa.ac.at Interim Report IR-12-071 Secondary sexual characteristics in codfishes (Gadidae) in relation to sound production, habitat use, and social behaviour Jon Egil Skjæraasen Justin J. -

Updated Checklist of Marine Fishes (Chordata: Craniata) from Portugal and the Proposed Extension of the Portuguese Continental Shelf
European Journal of Taxonomy 73: 1-73 ISSN 2118-9773 http://dx.doi.org/10.5852/ejt.2014.73 www.europeanjournaloftaxonomy.eu 2014 · Carneiro M. et al. This work is licensed under a Creative Commons Attribution 3.0 License. Monograph urn:lsid:zoobank.org:pub:9A5F217D-8E7B-448A-9CAB-2CCC9CC6F857 Updated checklist of marine fishes (Chordata: Craniata) from Portugal and the proposed extension of the Portuguese continental shelf Miguel CARNEIRO1,5, Rogélia MARTINS2,6, Monica LANDI*,3,7 & Filipe O. COSTA4,8 1,2 DIV-RP (Modelling and Management Fishery Resources Division), Instituto Português do Mar e da Atmosfera, Av. Brasilia 1449-006 Lisboa, Portugal. E-mail: [email protected], [email protected] 3,4 CBMA (Centre of Molecular and Environmental Biology), Department of Biology, University of Minho, Campus de Gualtar, 4710-057 Braga, Portugal. E-mail: [email protected], [email protected] * corresponding author: [email protected] 5 urn:lsid:zoobank.org:author:90A98A50-327E-4648-9DCE-75709C7A2472 6 urn:lsid:zoobank.org:author:1EB6DE00-9E91-407C-B7C4-34F31F29FD88 7 urn:lsid:zoobank.org:author:6D3AC760-77F2-4CFA-B5C7-665CB07F4CEB 8 urn:lsid:zoobank.org:author:48E53CF3-71C8-403C-BECD-10B20B3C15B4 Abstract. The study of the Portuguese marine ichthyofauna has a long historical tradition, rooted back in the 18th Century. Here we present an annotated checklist of the marine fishes from Portuguese waters, including the area encompassed by the proposed extension of the Portuguese continental shelf and the Economic Exclusive Zone (EEZ). The list is based on historical literature records and taxon occurrence data obtained from natural history collections, together with new revisions and occurrences. -

Elasmobranch Biodiversity, Conservation and Management Proceedings of the International Seminar and Workshop, Sabah, Malaysia, July 1997
The IUCN Species Survival Commission Elasmobranch Biodiversity, Conservation and Management Proceedings of the International Seminar and Workshop, Sabah, Malaysia, July 1997 Edited by Sarah L. Fowler, Tim M. Reed and Frances A. Dipper Occasional Paper of the IUCN Species Survival Commission No. 25 IUCN The World Conservation Union Donors to the SSC Conservation Communications Programme and Elasmobranch Biodiversity, Conservation and Management: Proceedings of the International Seminar and Workshop, Sabah, Malaysia, July 1997 The IUCN/Species Survival Commission is committed to communicate important species conservation information to natural resource managers, decision-makers and others whose actions affect the conservation of biodiversity. The SSC's Action Plans, Occasional Papers, newsletter Species and other publications are supported by a wide variety of generous donors including: The Sultanate of Oman established the Peter Scott IUCN/SSC Action Plan Fund in 1990. The Fund supports Action Plan development and implementation. To date, more than 80 grants have been made from the Fund to SSC Specialist Groups. The SSC is grateful to the Sultanate of Oman for its confidence in and support for species conservation worldwide. The Council of Agriculture (COA), Taiwan has awarded major grants to the SSC's Wildlife Trade Programme and Conservation Communications Programme. This support has enabled SSC to continue its valuable technical advisory service to the Parties to CITES as well as to the larger global conservation community. Among other responsibilities, the COA is in charge of matters concerning the designation and management of nature reserves, conservation of wildlife and their habitats, conservation of natural landscapes, coordination of law enforcement efforts as well as promotion of conservation education, research and international cooperation. -

Marine Fishes from Galicia (NW Spain): an Updated Checklist
1 2 Marine fishes from Galicia (NW Spain): an updated checklist 3 4 5 RAFAEL BAÑON1, DAVID VILLEGAS-RÍOS2, ALBERTO SERRANO3, 6 GONZALO MUCIENTES2,4 & JUAN CARLOS ARRONTE3 7 8 9 10 1 Servizo de Planificación, Dirección Xeral de Recursos Mariños, Consellería de Pesca 11 e Asuntos Marítimos, Rúa do Valiño 63-65, 15703 Santiago de Compostela, Spain. E- 12 mail: [email protected] 13 2 CSIC. Instituto de Investigaciones Marinas. Eduardo Cabello 6, 36208 Vigo 14 (Pontevedra), Spain. E-mail: [email protected] (D. V-R); [email protected] 15 (G.M.). 16 3 Instituto Español de Oceanografía, C.O. de Santander, Santander, Spain. E-mail: 17 [email protected] (A.S); [email protected] (J.-C. A). 18 4Centro Tecnológico del Mar, CETMAR. Eduardo Cabello s.n., 36208. Vigo 19 (Pontevedra), Spain. 20 21 Abstract 22 23 An annotated checklist of the marine fishes from Galician waters is presented. The list 24 is based on historical literature records and new revisions. The ichthyofauna list is 25 composed by 397 species very diversified in 2 superclass, 3 class, 35 orders, 139 1 1 families and 288 genus. The order Perciformes is the most diverse one with 37 families, 2 91 genus and 135 species. Gobiidae (19 species) and Sparidae (19 species) are the 3 richest families. Biogeographically, the Lusitanian group includes 203 species (51.1%), 4 followed by 149 species of the Atlantic (37.5%), then 28 of the Boreal (7.1%), and 17 5 of the African (4.3%) groups. We have recognized 41 new records, and 3 other records 6 have been identified as doubtful. -
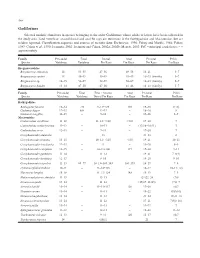
Gadiformes Selected Meristic Characters in Species Belonging to the Order Gadiformes Whose Adults Or Larvae Have Been Collected in the Study Area
548 Gadiformes Selected meristic characters in species belonging to the order Gadiformes whose adults or larvae have been collected in the study area. Total vertebrae, second dorsal and anal fin rays are numerous in the Bathygadidae and Macrouridae, but are seldom reported. Classification sequence and sources of meristic data: Eschmeyer, 1990; Fahay and Markle, 1984; Fahay, 1989; Cohen et al., 1990; Iwamoto, 2002; Iwamoto and Cohen, 2002a; 2002b; Merrett, 2003. PrC = principal caudal rays; ~ = approximately Family Precaudal Total Dorsal Anal Pectoral Pelvic Species Vertebrae Vertebrae Fin Rays Fin Rays Fin Rays Fin Rays Bregmacerotidae Bregmaceros atlanticus 14 53–55 47–56 49–58 16–21 5–7 Bregmaceros cantori 14 45–49 45–49 45–49 16–23 (family) 5–7 Bregmaceros sp. 14–15 52–59 52–59 58–69 16–23 (family) 5–7 Bregmaceros houdei 13–14 47–50 47–50 41–46 16–23 (family) 5–7 Family Precaudal Total First + Second Anal Pectoral Pelvic Species Vertebrae Vertebrae Dorsal Fin Rays Fin Rays Fin Rays Fin Rays Bathygadidae Bathygadus favosus 12–14 ~70 9–11+125 110 15–18 9(10) Gadomus dispar 12–13 80+ 12–13 – 18–20 8 Gadomus longifilis 11–13 – 9–11 – 14–16 8–9 Macrouridae Caelorinchus caribbeus 11–12 – 11–12+>110 >110 17–20 7 Caelorinchus coelorhynchus 11–12 – 10–11 – (17)18–20(21) 7 Caelorinchus occa 12–13 – 9–11 – 17–20 7 Coryphaenoides alateralis – 13 – 21–23 8 Coryphaenoides armatus 13–15 – 10–12+~125 ~135 19–21 10–11 Coryphaenoides brevibarbis 12–13 – 9 – 19–20 8–9 Coryphaenoides carapinus 12–15 – 10–11+100 117 17–20 9–11 Coryphaenoides guentheri -

Reducing the Conflict Between Cormorants and Fisheries on a Pan-European Scale
Reducing the conflict between Cormorants and fisheries on a pan-European scale REDCAFE Final Report Report of a Concerted Action funded by the European Union. Study contract no. Q5CA-2000-31387: Reducing the conflict between cormorants and fisheries on a pan-European scale. Edited by D N Carss Centre for Ecology & Hydrology Banchory, Hill of Brathens, Banchory Aberdeenshire, AB31 4BW, Scotland, UK. Natural Environment Research Council Centre for Ecology & Hydrology CEH Banchory Hill of Brathens Banchory Aberdeenshire AB31 4BY Scotland, UK. Fax: +44 1330 823303 Internet: [email protected] Web: www.banchory.ceh.ac.uk CEH Contract Number: C01749 Reducing the conflict between cormorants and fisheries on a pan-European scale Final Report Edited by: D N Carss Commissioned by: European Commission DG XIV Directorate-General for Fisheries Rue de la Loi 200 Bâtiment J II 99 6/11 B-1049 Brussels, Belgium Contract number: Q5CA-2000-31387 This report does not necessarily reflect the views of the European Commission and in no way anticipates any future opinion of the Commission. The contents of the report may not be reproduced unless the source of the material is indicated. This study has been carried out with the financial assistance of the European Commission. This report is drafted at the request of the commissioner indicated above and is his property. Nothing from this report may be reproduced and/or published by print, photoprint microfilm or any other means without the previous written consent from the commissioner of the study. Photographs on the cover (Top left, clockwise): Great cormorant © David Grémillet, coastal fisheries, Greece © Dave Carss, recreational fisherman © Trout & Salmon magazine, Danish pound net fishermen, © DC, Roach © Roger Phillips and Martyn Rix, REDCAFE participants, Waltham Abbey, November 2002, © Szymon Bzoma. -

Reproductive Potential of Pacific Cod (Gadus Macrocephalus) in Alaska
Reproductive Potential Of Pacific Cod (Gadus Macrocephalus) In Alaska Item Type Thesis Authors Ormseth, Olav Aleksander Download date 28/09/2021 04:53:02 Link to Item http://hdl.handle.net/11122/8941 REPRODUCTIVE POTENTIAL OF PACIFIC COD (GADUS MACROCEPHALUS) IN ALASKA A THESIS Presented to the Faculty of the University of Alaska Fairbanks in Partial Fulfillment of the Requirements for the Degree of DOCTOR OF PHILOSOPHY By Olav Aleksander Ormseth, M.S. Fairbanks, Alaska December 2007 Reproduced with permission of the copyright owner. Further reproduction prohibited without permission. UMI Number: 3302509 INFORMATION TO USERS The quality of this reproduction is dependent upon the quality of the copy submitted. Broken or indistinct print, colored or poor quality illustrations and photographs, print bleed-through, substandard margins, and improper alignment can adversely affect reproduction. In the unlikely event that the author did not send a complete manuscript and there are missing pages, these will be noted. Also, if unauthorized copyright material had to be removed, a note will indicate the deletion. ® UMI UMI Microform 3302509 Copyright 2008 by ProQuest LLC. All rights reserved. This microform edition is protected against unauthorized copying under Title 17, United States Code. ProQuest LLC 789 E. Eisenhower Parkway PO Box 1346 Ann Arbor, Ml 48106-1346 Reproduced with permission of the copyright owner. Further reproduction prohibited without permission. REPRODUCTIVE POTENTIAL OF PACIFIC COD (GADUS MACROCEPHALUS) IN ALASKA By Olav Aleksander Ormseth RECOMMENDED: A Advisory Committee Chair Head, Program in Marine Science and Limnology APPROVED: DearTT^chool of Fisheries and Oc&an Sciences Dean of the Graduate $ 'f - 2 o 0 7 Date Reproduced with permission of the copyright owner. -

The Complete Mitochondrial Genome of the Whiting, Merlangius
CORE Metadata, citation and similar papers at core.ac.uk Provided by Digital.CSIC The complete mitochondrial genome of the whiting, Merlangius merlangus and the haddock, Melanogrammus aeglefinus: A detailed genomic comparison among closely related species of the Gadidae family Severine Roques a , Clive J. Fox b , Maria I. Villasana a , Ciro Rico a,⁎ a Estación Biológica de Doñana, CSIC, Pabellón del Perú, 41013 Sevilla, Spain b The Centre for Environment, Fisheries and Aquaculture Science, Pakefield Road, Lowestoft, Suffolk, NR33 OHT, England, United Kingdom Abstract We determined the first complete mitochondrial DNA (mtDNA) sequences for the whiting (Merlangius merlangus, family Gadidae, order Gadiformes) and the haddock (Melanogrammus aeglefinus, family Gadidae, order Gadiformes). The entire mitogenomes were amplified and sequenced by primer walking using newly designed specific internal primers. Lengths were 16,569 and 16,585 bases for whiting and haddock respectively, lengths which lie within the range of previously reported gadiform sequences from Atlantic cod (Gadus morhua, 16,696 bases) and walleye pollock (Theragra chalcogramma, 16,570 bases). Gene arrangement in both species conformed to the order seen in most vertebrate mitochondrial genomes. We identified a long intergenic spacer located between the tRNAThr and tRNAPro genes (of 100 and 70 bp long for whiting and haddock, respectively), as previously described for other species of the order Gadiformes. Using nucleotide and amino acid divergence data of four complete gadoid mitogenomes (M. merlangius, M. aeglefinus, G. morhua and T. chalcogramma), we examined in detail the relative mtDNA mutation patterns across genes and among Gadidae species and tested for the performance of each protein-coding, transfer RNA and ribosomal RNA gene in depicting the expected phylogeny among the four species, as compared with the whole genome dataset. -

PDF Download
An illustrated key to the described valid species of Trimma (Teleostei: Gobiidae) RICHARD WINTERBOTTOM Curator Emeritus, Department of Natural History, Royal Ontario Museum, 100 Queen’s Park, Toronto, Ontario, M5S 2C6, Canada Professor Emeritus, Department of Ecology & Evolutionary Biology, University of Toronto, Toronto, Ontario, M5S 1A1, Canada E-mail: [email protected] Abstract An illustrated key to the 105 recognized valid species of the gobiid Trimma (pygmygobies), based on morphological and color-pattern characteristics is presented. Caution should be employed when using the key for specimens less than about 16 mm SL. A brief summary of what we know about the biology of the various species is given. Definitions and illustrations of the various characters used in the key are provided, along with the geographic range (where discernible) for each species. Details of the catalogue number and provenance of the holotype of each species (where known) are included. Comments on the variation of the mitochondrial genetic marker COI- 5P, derived from an unpublished neighbor-joining network based on 844 tissue samples, are also included where available. Key words: ichthyology, taxonomy, coral-reef fishes, pygymygobies, Indo-Pacific Ocean, mtDNA, barcoding. Citation: Winterbottom, R. (2019) An illustrated key to the described valid species of Trimma (Teleostei: Gobiidae). Journal of the Ocean Science Foundation, 34, 1–61. doi: https://doi.org/10.5281/zenodo.3525430 Introduction In 1906, Jordan & Seale erected a new genus, Trimma, for a new species they described from Samoa, T. caesiura. The diagnosis of their new taxon is not especially enlightening. They merely stated that it was similar to Eviota, but differed in having the region before the dorsal fin fully scaled, and that the “texture” of “the species is firmer and less translucent” (op.cit., p. -

Length-Weight Relationships of Marine Fish Collected from Around the British Isles
Science Series Technical Report no. 150 Length-weight relationships of marine fish collected from around the British Isles J. F. Silva, J. R. Ellis and R. A. Ayers Science Series Technical Report no. 150 Length-weight relationships of marine fish collected from around the British Isles J. F. Silva, J. R. Ellis and R. A. Ayers This report should be cited as: Silva J. F., Ellis J. R. and Ayers R. A. 2013. Length-weight relationships of marine fish collected from around the British Isles. Sci. Ser. Tech. Rep., Cefas Lowestoft, 150: 109 pp. Additional copies can be obtained from Cefas by e-mailing a request to [email protected] or downloading from the Cefas website www.cefas.defra.gov.uk. © Crown copyright, 2013 This publication (excluding the logos) may be re-used free of charge in any format or medium for research for non-commercial purposes, private study or for internal circulation within an organisation. This is subject to it being re-used accurately and not used in a misleading context. The material must be acknowledged as Crown copyright and the title of the publication specified. This publication is also available at www.cefas.defra.gov.uk For any other use of this material please apply for a Click-Use Licence for core material at www.hmso.gov.uk/copyright/licences/ core/core_licence.htm, or by writing to: HMSO’s Licensing Division St Clements House 2-16 Colegate Norwich NR3 1BQ Fax: 01603 723000 E-mail: [email protected] 3 Contents Contents 1. Introduction 5 2. -

Establishment and Partial Characterization of a Cell Line from Burbot Lota Lota Maculosa: Susceptibility to IHNV, IPNV and VHSV Mark P
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln USGS Staff -- ubP lished Research US Geological Survey 5-18-2010 Establishment and partial characterization of a cell line from burbot Lota lota maculosa: susceptibility to IHNV, IPNV and VHSV Mark P. Polinski Department of Fish and Wildlife Resources, College of Natural Resources, University of Idaho John D. Drennan Intervet/Schering-Plough Animal Health William N. Batts US Geological Survey, Western Fisheries Research Center Susan C. Ireland Kootenai Tribe of Idaho Kenneth D. Cain Department of Fish and Wildlife Resources, College of Natural Resources, University of Idaho,, [email protected] Follow this and additional works at: http://digitalcommons.unl.edu/usgsstaffpub Part of the Geology Commons, Oceanography and Atmospheric Sciences and Meteorology Commons, Other Earth Sciences Commons, and the Other Environmental Sciences Commons Polinski, Mark P.; Drennan, John D.; Batts, iW lliam N.; Ireland, Susan C.; and Cain, Kenneth D., "Establishment and partial characterization of a cell line from burbot Lota lota maculosa: susceptibility to IHNV, IPNV and VHSV" (2010). USGS Staff -- Published Research. 1062. http://digitalcommons.unl.edu/usgsstaffpub/1062 This Article is brought to you for free and open access by the US Geological Survey at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in USGS Staff -- ubP lished Research by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Vol. 90: 15–23, 2010 DISEASES OF AQUATIC ORGANISMS Published May 18 doi: 10.3354/dao02215 Dis Aquat Org Establishment and partial characterization of a cell line from burbot Lota lota maculosa: susceptibility to IHNV, IPNV and VHSV Mark P.