Supplemental Table II. Molecular Network Components of Genotype-Independent Stress Response
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Metabolic Engineering of Microbial Cell Factories for Biosynthesis of Flavonoids: a Review
molecules Review Metabolic Engineering of Microbial Cell Factories for Biosynthesis of Flavonoids: A Review Hanghang Lou 1,†, Lifei Hu 2,†, Hongyun Lu 1, Tianyu Wei 1 and Qihe Chen 1,* 1 Department of Food Science and Nutrition, Zhejiang University, Hangzhou 310058, China; [email protected] (H.L.); [email protected] (H.L.); [email protected] (T.W.) 2 Hubei Key Lab of Quality and Safety of Traditional Chinese Medicine & Health Food, Huangshi 435100, China; [email protected] * Correspondence: [email protected]; Tel.: +86-0571-8698-4316 † These authors are equally to this manuscript. Abstract: Flavonoids belong to a class of plant secondary metabolites that have a polyphenol structure. Flavonoids show extensive biological activity, such as antioxidative, anti-inflammatory, anti-mutagenic, anti-cancer, and antibacterial properties, so they are widely used in the food, phar- maceutical, and nutraceutical industries. However, traditional sources of flavonoids are no longer sufficient to meet current demands. In recent years, with the clarification of the biosynthetic pathway of flavonoids and the development of synthetic biology, it has become possible to use synthetic metabolic engineering methods with microorganisms as hosts to produce flavonoids. This article mainly reviews the biosynthetic pathways of flavonoids and the development of microbial expression systems for the production of flavonoids in order to provide a useful reference for further research on synthetic metabolic engineering of flavonoids. Meanwhile, the application of co-culture systems in the biosynthesis of flavonoids is emphasized in this review. Citation: Lou, H.; Hu, L.; Lu, H.; Wei, Keywords: flavonoids; metabolic engineering; co-culture system; biosynthesis; microbial cell factories T.; Chen, Q. -

Key Enzymes Involved in the Synthesis of Hops Phytochemical Compounds: from Structure, Functions to Applications
International Journal of Molecular Sciences Review Key Enzymes Involved in the Synthesis of Hops Phytochemical Compounds: From Structure, Functions to Applications Kai Hong , Limin Wang, Agbaka Johnpaul , Chenyan Lv * and Changwei Ma * College of Food Science and Nutritional Engineering, China Agricultural University, 17 Qinghua Donglu Road, Haidian District, Beijing 100083, China; [email protected] (K.H.); [email protected] (L.W.); [email protected] (A.J.) * Correspondence: [email protected] (C.L.); [email protected] (C.M.); Tel./Fax: +86-10-62737643 (C.M.) Abstract: Humulus lupulus L. is an essential source of aroma compounds, hop bitter acids, and xanthohumol derivatives mainly exploited as flavourings in beer brewing and with demonstrated potential for the treatment of certain diseases. To acquire a comprehensive understanding of the biosynthesis of these compounds, the primary enzymes involved in the three major pathways of hops’ phytochemical composition are herein critically summarized. Hops’ phytochemical components impart bitterness, aroma, and antioxidant activity to beers. The biosynthesis pathways have been extensively studied and enzymes play essential roles in the processes. Here, we introduced the enzymes involved in the biosynthesis of hop bitter acids, monoterpenes and xanthohumol deriva- tives, including the branched-chain aminotransferase (BCAT), branched-chain keto-acid dehydroge- nase (BCKDH), carboxyl CoA ligase (CCL), valerophenone synthase (VPS), prenyltransferase (PT), 1-deoxyxylulose-5-phosphate synthase (DXS), 4-hydroxy-3-methylbut-2-enyl diphosphate reductase (HDR), Geranyl diphosphate synthase (GPPS), monoterpene synthase enzymes (MTS), cinnamate Citation: Hong, K.; Wang, L.; 4-hydroxylase (C4H), chalcone synthase (CHS_H1), chalcone isomerase (CHI)-like proteins (CHIL), Johnpaul, A.; Lv, C.; Ma, C. -

Building Blue E. Coli: Engineering the Anthocyanin Biosynthetic Pathway
Building blue E. coli: Engineering the anthocyanin biosynthetic pathway Leah Johnston1, Lucas Jarche1, Alexander Porter1, Julie Ryu1, Emma Price1, Emma Finlayson-Trick1, Khaled Aly1, Paul Bissonnette2, Eric Pace3, Dr. John Rohde1 1Department of Microbiology and Immunology, 2Department of Chemistry, 3Department of Agriculture Dalhousie University, Halifax, Canada, B3H 1X5 THE ANTHOCYANIN PATHWAY METHODS RESULTS 1kb (B) Method of Template DNA for each of the genes required for the production of the Anthocyanin biosynthetic Gene Size (bp) Function Epitope ladder (B) Acquisition of pathway in E. coli were acquired from the following sources outlined in Figure 1 (B). (A) Name Tag Template DNA 10 Phenylalanine Lyase involved in Amplified from A. (A) 2762 polyphenol S-Tag thaliana cDNA, Ammonia . synthesis. insert in Phenylalanine Ammonia Lyase and 4-Coumaroyl CoA Ligase were acquired as inserts in a 3.0. Lyase pACYCDuet-1 pACYCDuet-1 plasmid from Sinyuan Wang et. al (2005) at Utah State University. Cinnamate 4- 2.0 Phenylalanine from Utah State Hydroxylase-HA fusion protein was inserted into this plasmid at the end of the PAL coding region 1.5 Phenylalanine ammonia lyase University using restriction enzymes and transformed into BL21 CaCl2 competent E. coli with T7 RNA Cinnamate 4- Oxidoreductase Amplified from A. Cinnamic Acid 1518 involved in HA-Tag thaliana cDNA, Polymerase activity. A western blot with anti-HA antibody was used to detect the presence of the 1.0 Hydroxylase phenylalanine purchased from C4H fusion protein in the resultant cells. Cinnamate 4-hydroxylase metabolism. TAIR database P-Coumaric Acid 4-Coumaroyl Ligase involved in Insert in 2546 phenylpropanoid His-Tag pACYCDuet-1 Chalcone Synthase, Chalcone Isomerase and Flavanone 3-Hydroxylase were amplified from the CoA Ligase 0.5 4-coumaryl-coenzyme A ligase biosynthesis. -

Analysis of the Chromosomal Clustering of Fusarium-Responsive
www.nature.com/scientificreports OPEN Analysis of the chromosomal clustering of Fusarium‑responsive wheat genes uncovers new players in the defence against head blight disease Alexandre Perochon1,2, Harriet R. Benbow1,2, Katarzyna Ślęczka‑Brady1, Keshav B. Malla1 & Fiona M. Doohan1* There is increasing evidence that some functionally related, co‑expressed genes cluster within eukaryotic genomes. We present a novel pipeline that delineates such eukaryotic gene clusters. Using this tool for bread wheat, we uncovered 44 clusters of genes that are responsive to the fungal pathogen Fusarium graminearum. As expected, these Fusarium‑responsive gene clusters (FRGCs) included metabolic gene clusters, many of which are associated with disease resistance, but hitherto not described for wheat. However, the majority of the FRGCs are non‑metabolic, many of which contain clusters of paralogues, including those implicated in plant disease responses, such as glutathione transferases, MAP kinases, and germin‑like proteins. 20 of the FRGCs encode nonhomologous, non‑metabolic genes (including defence‑related genes). One of these clusters includes the characterised Fusarium resistance orphan gene, TaFROG. Eight of the FRGCs map within 6 FHB resistance loci. One small QTL on chromosome 7D (4.7 Mb) encodes eight Fusarium‑ responsive genes, fve of which are within a FRGC. This study provides a new tool to identify genomic regions enriched in genes responsive to specifc traits of interest and applied herein it highlighted gene families, genetic loci and biological pathways of importance in the response of wheat to disease. Prokaryote genomes encode co-transcribed genes with related functions that cluster together within operons. Clusters of functionally related genes also exist in eukaryote genomes, including fungi, nematodes, mammals and plants1. -

Diversity of Plant Virus Movement Proteins: What Do They Have in Common?
processes Review Diversity of Plant Virus Movement Proteins: What Do They Have in Common? Yuri L. Dorokhov 1,2,* , Ekaterina V. Sheshukova 1, Tatiana E. Byalik 3 and Tatiana V. Komarova 1,2 1 Vavilov Institute of General Genetics Russian Academy of Sciences, 119991 Moscow, Russia; [email protected] (E.V.S.); [email protected] (T.V.K.) 2 Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119991 Moscow, Russia 3 Department of Oncology, I.M. Sechenov First Moscow State Medical University, 119991 Moscow, Russia; [email protected] * Correspondence: [email protected] Received: 11 November 2020; Accepted: 24 November 2020; Published: 26 November 2020 Abstract: The modern view of the mechanism of intercellular movement of viruses is based largely on data from the study of the tobacco mosaic virus (TMV) 30-kDa movement protein (MP). The discovered properties and abilities of TMV MP, namely, (a) in vitro binding of single-stranded RNA in a non-sequence-specific manner, (b) participation in the intracellular trafficking of genomic RNA to the plasmodesmata (Pd), and (c) localization in Pd and enhancement of Pd permeability, have been used as a reference in the search and analysis of candidate proteins from other plant viruses. Nevertheless, although almost four decades have passed since the introduction of the term “movement protein” into scientific circulation, the mechanism underlying its function remains unclear. It is unclear why, despite the absence of homology, different MPs are able to functionally replace each other in trans-complementation tests. Here, we consider the complexity and contradictions of the approaches for assessment of the ability of plant viral proteins to perform their movement function. -

Supplementary Material
CSIRO PUBLISHING www.publish.csiro.au/journals/fpb Functional Plant Biology 33, 1–24 Supplementary material: FP05139_AC Supplementary material Molecular targets of elevated [CO2] in leaves and stems of Populus deltoides: implications for future tree growth and carbon sequestration Nathalie DruartA,B, Marisa Rodríguez-BueyA, Greg Barron-GaffordC, Andreas SjödinA, Rishikesh BhaleraoB and Vaughan HurryA,D AUmeå Plant Science Centre, Department of Plant Physiology, Umeå University, S-901 87 Umeå, Sweden. BUmeå Plant Science Centre, Department of Forest Genetics and Plant Physiology, Swedish University of Agricultural Sciences, S-901 83 Umeå, Sweden. CBiosphere 2 Laboratory, Columbia University, Oracle AZ 85623 USA. Current address: Ecology and Evolutionary Biology, University of Arizona, Tucson, AZ 85719, USA. DCorresponding author. Email: [email protected] Appendix S1 1.0 1.0 A B 0.5 0.5 0.0 0.0 –0.5 –0.5 –1.0 –1.0 Relative gene expression (log gene expression Relative ) 1.0 1.0 2 C D 0.5 0.5 0.0 0.0 –0.5 –0.5 2 ) Relative gene expression (log gene expression Relative –1.0 –1.0 1.0 1.0 E F 0.5 0.5 0.0 0.0 –0.5 –0.5 –1.0 –1.0 400 800 1200 400 800 1200 Ambient [CO2] (µmol mol–1) Validation of reproducibility of the hybridisations by comparing the hybridisation results of three genes for which two independent cDNA clones had been spotted on the array. (A, C, E leaves; B, D, F stems). (A, B) Protein phosphatase 2A, 65 kDa subunit. (C, D) Elicitor-inducible cytochrome P450. -
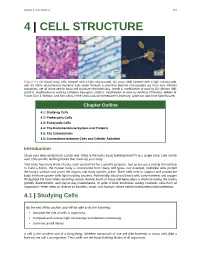
Chapter 4 – Cell Structure
Chapter 4 | Cell Structure 107 4 | CELL STRUCTURE Figure 4.1 (a) Nasal sinus cells (viewed with a light microscope), (b) onion cells (viewed with a light microscope), and (c) Vibrio tasmaniensis bacterial cells (seen through a scanning electron microscope) are from very different organisms, yet all share certain basic cell structure characteristics. (credit a: modification of work by Ed Uthman, MD; credit b: modification of work by Umberto Salvagnin; credit c: modification of work by Anthony D'Onofrio, William H. Fowle, Eric J. Stewart, and Kim Lewis of the Lewis Lab at Northeastern University; scale-bar data from Matt Russell) Chapter Outline 4.1: Studying Cells 4.2: Prokaryotic Cells 4.3: Eukaryotic Cells 4.4: The Endomembrane System and Proteins 4.5: The Cytoskeleton 4.6: Connections between Cells and Cellular Activities Introduction Close your eyes and picture a brick wall. What is the wall's basic building block? It is a single brick. Like a brick wall, cells are the building blocks that make up your body. Your body has many kinds of cells, each specialized for a specific purpose. Just as we use a variety of materials to build a home, the human body is constructed from many cell types. For example, epithelial cells protect the body's surface and cover the organs and body cavities within. Bone cells help to support and protect the body. Immune system cells fight invading bacteria. Additionally, blood and blood cells carry nutrients and oxygen throughout the body while removing carbon dioxide. Each of these cell types plays a vital role during the body's growth, development, and day-to-day maintenance. -

Development and Function of Plasmodesmata in Zygotes of Fucus Distichus
Botanica Marina 2015; 58(3): 229–238 Chikako Nagasato*, Makoto Terauchi, Atsuko Tanaka and Taizo Motomura Development and function of plasmodesmata in zygotes of Fucus distichus Abstract: Brown algae have plasmodesmata, tiny tubu- Introduction lar cytoplasmic channels connecting adjacent cells. The lumen of plasmodesmata is 10–20 nm wide, and it takes a Multicellular organisms such as animals, fungi, land simple form, without a desmotubule (the inner membrane plants, and brown algae have specific cellular connections. structure consisting of endoplasmic reticulum in the plas- These structures connect the cytoplasm of adjacent cells modesmata of green plants). In this study, we analyzed and provide a route for cell-cell communication. Green the ultrastructure and distribution of plasmodesmata plants, including land plants and certain species of green during development of Fucus distichus zygotes. The first algae (reviewed in Robards and Lucas 1990, Raven 1997), cytokinesis of zygotes in brown algae is not accompanied and brown algae (Bisalputra 1966) possess plasmodes- by plasmodesmata formation. As the germlings develop, mata. These are cytoplasmic canals that pass through the plasmodesmata are found in all septal cell walls, includ- septal cell wall and connect adjacent cells. Plasmodes- ing the first cell division plane. Plasmodesmata are formed mata in green plants and brown algae share similar char- de novo on the existing cell wall. Pit fields, which are clus- acteristics; however, plasmodesmata in brown algae lack ters of plasmodesmata, were observed in germlings with a desmotubule, a tubular strand of connecting endoplas- differentiated cell layers. Apart from the normal plas- mic reticulum (ER) that penetrates the plasmodesmata modesmata, these pit fields had branched plasmodes- of land plants (Terauchi et al. -

Transcription Factors Regulating Terpene Synthases in Tomato Trichomes
UvA-DARE (Digital Academic Repository) Transcription factors regulating terpene synthases in tomato trichomes Spyropoulou, E. Publication date 2012 Document Version Final published version Link to publication Citation for published version (APA): Spyropoulou, E. (2012). Transcription factors regulating terpene synthases in tomato trichomes. General rights It is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), other than for strictly personal, individual use, unless the work is under an open content license (like Creative Commons). Disclaimer/Complaints regulations If you believe that digital publication of certain material infringes any of your rights or (privacy) interests, please let the Library know, stating your reasons. In case of a legitimate complaint, the Library will make the material inaccessible and/or remove it from the website. Please Ask the Library: https://uba.uva.nl/en/contact, or a letter to: Library of the University of Amsterdam, Secretariat, Singel 425, 1012 WP Amsterdam, The Netherlands. You will be contacted as soon as possible. UvA-DARE is a service provided by the library of the University of Amsterdam (https://dare.uva.nl) Download date:03 Oct 2021 Transcription factors regulating terpene synthases in tomato trichomes factors regulating Transcription Transcription factors regulating terpene synthases in tomato trichomes Eleni Spyropoulou Eleni Spyropoulou Transcription factors regulating terpene synthases in tomato trichomes ACADEMISCH PROEFSCHRIFT ter verkrijging van de graad van doctor aan de Universiteit van Amsterdam op gezag van de Rector Magnificus prof. dr. D.C. van den Boom ten overstaan van een door het college voor promoties ingestelde commissie, in het openbaar te verdedigen in de Agnietenkapel op dinsdag 03 juli 2012, te 16:00 uur door Eleni Spyropoulou geboren te Marousi, Griekenland PROMOTIECOMISSIE Promotor Prof. -

Noncatalytic Chalcone Isomerase-Fold Proteins in Humulus Lupulus Are Auxiliary Components in Prenylated Flavonoid Biosynthesis
Noncatalytic chalcone isomerase-fold proteins in Humulus lupulus are auxiliary components in prenylated flavonoid biosynthesis Zhaonan Bana,b, Hao Qina, Andrew J. Mitchellc, Baoxiu Liua, Fengxia Zhanga, Jing-Ke Wengc,d, Richard A. Dixone,f,1, and Guodong Wanga,1 aState Key Laboratory of Plant Genomics and National Center for Plant Gene Research, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, 100101 Beijing, China; bUniversity of Chinese Academy of Sciences, 100049 Beijing, China; cWhitehead Institute for Biomedical Research, Cambridge, MA 02142; dDepartment of Biology, Massachusetts Institute of Technology, Cambridge, MA 02139; eBioDiscovery Institute, University of North Texas, Denton, TX 76203; and fDepartment of Biological Sciences, University of North Texas, Denton, TX 76203 Contributed by Richard A. Dixon, April 25, 2018 (sent for review February 6, 2018; reviewed by Joerg Bohlmann and Mattheos A. G. Koffas) Xanthohumol (XN) and demethylxanthohumol (DMX) are special- braries have been deposited in the TrichOME database [www. ized prenylated chalconoids with multiple pharmaceutical appli- planttrichome.org (18)], and numerous large RNAseq datasets from cations that accumulate to high levels in the glandular trichomes different hop tissues or cultivars have also been made publically of hops (Humulus lupulus L.). Although all structural enzymes in available. By mining the hops transcriptome data, we and others have the XN pathway have been functionally identified, biochemical functionally identified several key terpenophenolic biosynthetic en- mechanisms underlying highly efficient production of XN have zymes from hop glandular trichomes (1, 18–23); these include car- not been fully resolved. In this study, we characterized two non- boxyl CoA ligase (CCL) genes and two aromatic prenyltransferase catalytic chalcone isomerase (CHI)-like proteins (designated as (PT) genes (HlPT1L and HlPT2) (22, 23). -

De Novo Sequencing and Comparative Analysis of Holy and Sweet Basil
Rastogi et al. BMC Genomics 2014, 15:588 http://www.biomedcentral.com/1471-2164/15/588 RESEARCH ARTICLE Open Access De novo sequencing and comparative analysis of holy and sweet basil transcriptomes Shubhra Rastogi1, Seema Meena1, Ankita Bhattacharya1, Sumit Ghosh1, Rakesh Kumar Shukla1, Neelam Singh Sangwan2, Raj Kishori Lal3, Madan Mohan Gupta4, Umesh Chandra Lavania3, Vikrant Gupta1, Dinesh A Nagegowda1* and Ajit Kumar Shasany1* Abstract Background: Ocimum L. of family Lamiaceae is a well known genus for its ethnobotanical, medicinal and aromatic properties, which are attributed to innumerable phenylpropanoid and terpenoid compounds produced by the plant. To enrich genomic resources for understanding various pathways, de novo transcriptome sequencing of two important species, O. sanctum and O. basilicum, was carried out by Illumina paired-end sequencing. Results: The sequence assembly resulted in 69117 and 130043 transcripts with an average length of 1646 ± 1210.1 bp and 1363 ± 1139.3 bp for O. sanctum and O. basilicum, respectively. Out of the total transcripts, 59648 (86.30%) and 105470 (81.10%) from O. sanctum and O. basilicum, and respectively were annotated by uniprot blastx against Arabidopsis, rice and lamiaceae. KEGG analysis identified 501 and 952 transcripts from O. sanctum and O. basilicum, respectively, related to secondary metabolism with higher percentage of transcripts for biosynthesis of terpenoids in O. sanctum and phenylpropanoids in O. basilicum. Higher digital gene expression in O. basilicum was validated through qPCR and correlated to higher essential oil content and chromosome number (O. sanctum, 2n = 16; and O. basilicum, 2n = 48). Several CYP450 (26) and TF (40) families were identified having probable roles in primary and secondary metabolism. -

Comprehensive Metabolic and Transcriptomic Profiling of Various
www.nature.com/scientificreports OPEN Comprehensive metabolic and transcriptomic profling of various tissues provide insights for saponin Received: 17 October 2017 Accepted: 4 June 2018 biosynthesis in the medicinally Published: xx xx xxxx important Asparagus racemosus Prabhakar Lal Srivastava1, Anurag Shukla2 & Raviraj M. Kalunke2 Asparagus racemosus (Shatavari), belongs to the family Asparagaceae and is known as a “curer of hundred diseases” since ancient time. This plant has been exploited as a food supplement to enhance immune system and regarded as a highly valued medicinal plant in Ayurvedic medicine system for the treatment of various ailments such as gastric ulcers, dyspepsia, cardiovascular diseases, neurodegenerative diseases, cancer, as a galactogogue and against several other diseases. In depth metabolic fngerprinting of various parts of the plant led to the identifcation of 13 monoterpenoids exclusively present in roots. LC-MS profling led to the identifcation of a signifcant number of steroidal saponins (33). However, we have also identifed 16 triterpene saponins for the frst time in A. racemosus. In order to understand the molecular basis of biosynthesis of major components, transcriptome sequencing from three diferent tissues (root, leaf and fruit) was carried out. Functional annotation of A. racemosus transcriptome resulted in the identifcation of 153 transcripts involved in steroidal saponin biosynthesis, 45 transcripts in triterpene saponin biosynthesis, 44 transcripts in monoterpenoid biosynthesis and 79 transcripts in favonoid biosynthesis. These fndings will pave the way for better understanding of the molecular basis of steroidal saponin, triterpene saponin, monoterpenoids and favonoid biosynthesis in A. racemosus. Asparagus racemosus is one of the most valuable medicinal plants, regarded as a “Queen of herbs” in Ayurvedic health system and has been used worldwide to cure various diseases1.