Cfdna Sequencing: Technological Approaches and Bioinformatic Issues
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Absolute Measurement of the Tissue Origins of Cell-Free DNA in the Healthy State and Following Paracetamol Overdose Danny Laurent1, Fiona Semple1, Philip J
Laurent et al. BMC Medical Genomics (2020) 13:60 https://doi.org/10.1186/s12920-020-0705-2 RESEARCH ARTICLE Open Access Absolute measurement of the tissue origins of cell-free DNA in the healthy state and following paracetamol overdose Danny Laurent1, Fiona Semple1, Philip J. Starkey Lewis2, Elaine Rose1, Holly A. Black1, Jennifer Coe1, Stuart J. Forbes2, Mark J. Arends3, James W. Dear4 and Timothy J. Aitman1* Abstract Background: Despite the emergence of cell-free DNA (cfDNA) as a clinical biomarker in cancer, the tissue origins of cfDNA in healthy individuals have to date been inferred only by indirect and relative measurement methods, such as tissue-specific methylation and nucleosomal profiling. Methods: We performed the first direct, absolute measurement of the tissue origins of cfDNA, using tissue-specific knockout mouse strains, in both healthy mice and following paracetamol (APAP) overdose. We then investigated the utility of total cfDNA and the percentage of liver-specific cfDNA as clinical biomarkers in patients presenting with APAP overdose. Results: Analysis of cfDNA from healthy tissue-specific knockout mice showed that cfDNA originates predominantly from white and red blood cell lineages, with minor contribution from hepatocytes, and no detectable contribution from skeletal and cardiac muscle. Following APAP overdose in mice, total plasma cfDNA and the percentage fraction originating from hepatocytes increased by ~ 100 and ~ 19-fold respectively. Total cfDNA increased by an average of more than 236-fold in clinical samples from APAP overdose patients with biochemical evidence of liver injury, and 18-fold in patients without biochemically apparent liver injury. Measurement of liver-specific cfDNA, using droplet digital PCR and methylation analysis, revealed that the contribution of liver to cfDNA was increased by an average of 175-fold in APAP overdose patients with biochemically apparent liver injury compared to healthy subjects, but was not increased in overdose patients with normal liver function tests. -
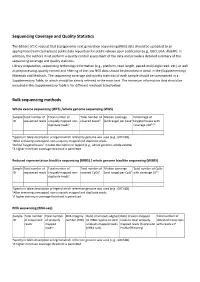
IJC Sequencing Coverage and Quality Statistics
Sequencing Coverage and Quality Statistics The Editors of IJC request that (epi)genomic next generation sequencing (NGS) data should be uploaded to an appropriate (restricted access) public data repository for public release upon publication (e.g., GEO, EGA, dbGAP). In addition, the authors must perform a quality control assessment of the data and provide a detailed summary of the sequencing coverage and quality statistics. Library preparation, sequencing technology information (e.g., platform, read length, paired‐end/single read, etc.) as well as preprocessing, quality control and filtering of the raw NGS data should be described in detail in the (Supplementary) Materials and Methods. The sequencing coverage and quality statistics of each sample should be summarized in a Supplementary Table, to which should be clearly referred in the main text. The minimum information that should be included in this Supplementary Table is for different methods listed below. Bulk sequencing methods Whole exome sequencing (WES) /whole genome sequencing (WGS) Sample Total number of Total number of Total number of Median coverage Percentage of ID sequenced reads uniquely mapped non‐ covered basesb (and range) per baseb targeted bases with duplicate readsa coverage ≥10b,c,d aSpecify in table description or legend which reference genome was used (e.g., GRCh38). bAfter removing unmapped, non‐uniquely mapped and duplicate reads. cDefine “targeted bases” in table description or legend (e.g., whole genome, whole exome). dA higher minimum coverage threshold is permitted. Reduced representation bisulfite sequencing (RRBS) / whole genome bisulfite sequencing (WGBS) Sample Total number of Total number of Total number of Median coverage Total number of CpGs ID sequenced reads uniquely mapped non‐ covered CpGsb (and range) per CpGb with coverage ≥5b,c duplicate readsa aSpecify in table description or legend which reference genome was used (e.g., GRCh38). -

MLH1 Single-Nucleotide Variant in Circulating Tumor DNA
www.nature.com/scientificreports OPEN MLH1 single‑nucleotide variant in circulating tumor DNA predicts overall survival of patients with hepatocellular carcinoma Soon Sun Kim1,4, Jung Woo Eun1,4, Ji‑Hye Choi2, Hyun Goo Woo2, Hyo Jung Cho1, Hye Ri Ahn3, Chul Won Suh3, Geum Ok Baek1, Sung Won Cho1 & Jae Youn Cheong1* Liquid biopsy can provide a strong basis for precision medicine. We aimed to identify novel single‑ nucleotide variants (SNVs) in circulating tumor DNA (ctDNA) in patients with hepatocellular carcinoma (HCC). Deep sequencing of plasma‑derived ctDNA from 59 patients with HCC was performed using a panel of 2924 SNVs in 69 genes. In 55.9% of the patients, at least one somatic mutation was detected. Among 25 SNVs in 12 genes, four frequently observed SNVs, MLH1 (13%), STK11 (13%), PTEN (9%), and CTNNB1 (4%), were validated using droplet digital polymerase chain reaction with ctDNA from 62 patients with HCC. Three candidate SNVs were detected in 35.5% of the patients, with a frequency of 19% for MLH1 chr3:37025749T>A, 11% for STK11 chr19:1223126C>G, and 8% for PTEN chr10:87864461C>G. The MLH1 and STK11 SNVs were also confrmed in HCC tissues. The presence of the MLH1 SNV, in combination with an increased ctDNA level, predicted poor overall survival among 107 patients. MLH1 chr3:37025749T>A SNV detection in ctDNA is feasible, and thus, ctDNA can be used to detect somatic mutations in HCC. Furthermore, the presence or absence of the MLH1 SNV in ctDNA, combined with the ctDNA level, can predict the prognosis of patients with HCC. -

Cell-Free DNA Next-Generation Sequencing in Pancreatobiliary Carcinomas
Published OnlineFirst June 24, 2015; DOI: 10.1158/2159-8290.CD-15-0274 ReseaRch BRief Cell-Free DNA Next-Generation Sequencing in Pancreatobiliary Carcinomas Oliver A. Zill1,2,3, Claire Greene2,4, Dragan Sebisanovic3, Lai Mun Siew3, Jim Leng2,4, Mary Vu5, Andrew E. Hendifar6, Zhen Wang2, Chloe E. Atreya2,4, Robin K. Kelley2,4, Katherine Van Loon2,4, Andrew H. Ko2,4, Margaret A. Tempero2,4, Trever G. Bivona2,4, Pamela N. Munster2,4, AmirAli Talasaz3, and Eric A. Collisson2,4 AbstRact Patients with pancreatic and biliary carcinomas lack personalized treatment options, in part because biopsies are often inadequate for molecular characteriza- tion. Cell-free DNA (cfDNA) sequencing may enable a precision oncology approach in this setting. We attempted to prospectively analyze 54 genes in tumor and cfDNA for 26 patients. Tumor sequencing failed in 9 patients (35%). In the remaining 17, 90.3% (95% confidence interval, 73.1%–97.5%) of mutations detected in tumor biopsies were also detected in cfDNA. The diagnostic accuracy of cfDNA sequencing was 97.7%, with 92.3% average sensitivity and 100% specificity across five informative genes. Changes in cfDNA correlated well with tumor marker dynamics in serial sampling (r = 0.93). We demonstrate that cfDNA sequencing is feasible, accurate, and sensitive in identifying tumor-derived mutations without prior knowledge of tumor genotype or the abundance of circulating tumor DNA. cfDNA sequencing should be considered in pancreatobiliary cancer trials where tissue sampling is unsafe, infeasible, or otherwise unsuccessful. SIGNIFICANCE: Precision medicine efforts in biliary and pancreatic cancers have been frustrated by difficulties in obtaining adequate tumor tissue for next-generation sequencing. -

Donor-Derived Cell-Free DNA in Kidney Transplantation As a Potential Rejection Biomarker: a Systematic Literature Review
Journal of Clinical Medicine Review Donor-Derived Cell-Free DNA in Kidney Transplantation as a Potential Rejection Biomarker: A Systematic Literature Review Adrian Martuszewski 1 , Patrycja Paluszkiewicz 1 , Magdalena Król 2, Mirosław Banasik 1 and Marta Kepinska 3,* 1 Department of Nephrology and Transplantation Medicine, Wroclaw Medical University, Borowska 213, 50-556 Wroclaw, Poland; [email protected] (A.M.); [email protected] (P.P.); [email protected] (M.B.) 2 Students Scientific Association, Department of Biomedical and Environmental Analysis, Faculty of Pharmacy, Wroclaw Medical University, 50-556 Wroclaw, Poland; [email protected] 3 Department of Biomedical and Environmental Analyses, Faculty of Pharmacy, Wroclaw Medical University, Borowska 211, 50-556 Wroclaw, Poland * Correspondence: [email protected]; Tel.: +48-71-784-0171 Abstract: Kidney transplantation (KTx) is the best treatment method for end-stage kidney disease. KTx improves the patient’s quality of life and prolongs their survival time; however, not all patients benefit fully from the transplantation procedure. For some patients, a problem is the premature loss of graft function due to immunological or non-immunological factors. Circulating cell-free DNA (cfDNA) is degraded deoxyribonucleic acid fragments that are released into the blood and other body fluids. Donor-derived cell-free DNA (dd-cfDNA) is cfDNA that is exogenous to the patient and comes from a transplanted organ. As opposed to an invasive biopsy, dd-cfDNA can be detected by a non-invasive analysis of a sample. The increase in dd-cfDNA concentration occurs even before the creatinine level starts rising, which may enable early diagnosis of transplant injury and adequate Citation: Martuszewski, A.; treatment to avoid premature graft loss. -

Circulating Tumor DNA As an Emerging Liquid Biopsy Biomarker
Int. J. Biol. Sci. 2020, Vol. 16 1551 Ivyspring International Publisher International Journal of Biological Sciences 2020; 16(9): 1551-1562. doi: 10.7150/ijbs.44024 Review Circulating tumor DNA as an emerging liquid biopsy biomarker for early diagnosis and therapeutic monitoring in hepatocellular carcinoma Xiaolin Wu1, Jiahui Li1, Asmae Gassa2, Denise Buchner1, Hakan Alakus1, Qiongzhu Dong3, Ning Ren4, Ming Liu5, Margarete Odenthal6, Dirk Stippel1, Christiane Bruns1, Yue Zhao1,7, and Roger Wahba1 1. Department of General, Visceral, Cancer and Transplantation Surgery, University Hospital of Cologne, Kerpener Straße 62, 50937, Cologne, Germany. 2. Department of Cardiothoracic Surgery, Heart Center, University Hospital of Cologne, Germany, Kerpener Straße 62, 5.937 Cologne, Germany. 3. Department of General Surgery, Huashan Hospital & Cancer Metastasis Institute & Institutes of Biomedical Sciences, Fudan University, 200032, Shanghai, P.R. China. 4. Liver Cancer Institute & Zhongshan Hospital; Department of Surgery, Institute of Fudan-Minhang Academic Health System, Minhang Branch, Zhongshan Hospital, Fudan University, 200032, Shanghai, P.R. China. 5. Affiliated Cancer Hospital & Institute of Guangzhou Medical University; Key Laboratory of Protein Modification and Degradation, School of Basic Medical Sciences, 510095, Guangzhou, P.R. China. 6. Institute of Pathology, University Hospital of Cologne, 50937, Cologne, Germany. 7. Department of General, Visceral und Vascular Surgery, Otto-von-Guericke University, 39120, Magdeburg, Germany. Corresponding authors: Roger Wahba ([email protected]; Tel: +49-221-478-4803); Yue Zhao (yue.zhao @uk-koeln.de; Tel: +49-221-478-30601). © The author(s). This is an open access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0/). -

Circulating Cell-Free Tumor DNA Analysis of 50 Genes by Next
Published OnlineFirst January 12, 2016; DOI: 10.1158/1078-0432.CCR-15-2470 Personalized Medicine and Imaging Clinical Cancer Research Circulating Cell-Free Tumor DNA Analysis of 50 Genes by Next-Generation Sequencing in the Prospective MOSCATO Trial Cecile Jovelet1, Ecaterina Ileana1,2, Marie-Cecile Le Deley3,4,5, Nelly Motte1, Silvia Rosellini3, Alfredo Romero1, Celine Lefebvre6, Marion Pedrero6,Noemie Pata-Merci7, Nathalie Droin7, Marc Deloger8, Christophe Massard2, Antoine Hollebecque2, Charles Ferte9, Amelie Boichard1, Sophie Postel-Vinay2,6, Maud Ngo-Camus2, Thierry De Baere10, Philippe Vielh11, Jean-Yves Scoazec1,5,11, Gilles Vassal12, Alexander Eggermont5,9, Fabrice Andre5,6,9, Jean-Charles Soria2,5,6, and Ludovic Lacroix1,6,11,13 Abstract Purpose: Liquid biopsies based on circulating cell-free DNA 48.4%–61.6%] at the patient level. Among the 220 patients with (cfDNA) analysis are described as surrogate samples for molecular mutations in tDNA, the sensitivity of cfDNA analysis was signif- analysis. We evaluated the concordance between tumor DNA icantly linked to the number of metastatic sites, albumin level, (tDNA) and cfDNA analysis on a large cohort of patients with tumor type, and number of lines of treatment. A sensitivity advanced or metastatic solid tumor, eligible for phase I trial and prediction score could be derived from clinical parameters. Sen- with good performance status, enrolled in MOSCATO 01 trial sitivity is 83% in patients with a high score (8). In addition, we (clinical trial NCT01566019). analyzed cfDNA for 51 patients without available tissue sample. Experimental Design: Blood samples were collected at inclu- Mutations were detected for 22 patients, including 19 oncogenic sion and cfDNA was extracted from plasma for 334 patients. -

Detection and Characterization of Low and High Genome Coverage Regions Using an Efficient Running Median and a Double Threshold Approach
bioRxiv preprint doi: https://doi.org/10.1101/092478; this version posted December 8, 2016. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under a CC-BY-NC 4.0 International license. Bioinformatics doi.10.1093/bioinformatics/xxxxxx Advance Access Publication Date: 2 April 2015 Applications Note Genome analysis Detection and characterization of low and high genome coverage regions using an efficient running median and a double threshold approach. Dimitri Desvillechabrol 1;∗, Christiane Bouchier 2, Sean Kennedy 1, and Thomas Cokelaer 3∗ 1Institut Pasteur – Pole Biomics 2Institut Pasteur – Genomic Platform – Pole Biomics 3Institut Pasteur – Bioinformatics and Biostatistics Hub – C3BI, USR 3756 IP CNRS – Paris, France ∗To whom correspondence should be addressed. Associate Editor: XXXXXXX Received on XXXXX; revised on XXXXX; accepted on XXXXX Abstract Motivation: Next Generation Sequencing (NGS) provides researchers with powerful tools to investigate both prokaryotic and eukaryotic genetics. An accurate assessment of reads mapped to a specific genome consists of inspecting the genome coverage as number of reads mapped to a specific genome location. Most current methods use the average of the genome coverage (sequencing depth) to summarize the overall coverage. This metric quickly assess the sequencing quality but ignores valuable biological information like the presence of repetitive regions or deleted genes. The detection of such information may be challenging due to a wide spectrum of heterogeneous coverage regions, a mixture of underlying models or the presence of a non-constant trend along the genome. -

Circulating Cell-Free DNA in Breast Cancer: Searching for Hidden Information Towards Precision Medicine
cancers Review Circulating Cell-Free DNA in Breast Cancer: Searching for Hidden Information towards Precision Medicine Maria Panagopoulou 1,*, Manel Esteller 2,3,4,5 and Ekaterini Chatzaki 1,6 1 Laboratory of Pharmacology, Department of Medicine, Democritus University of Thrace (DUTH), 68100 Alexandroupolis, Greece; [email protected] 2 Josep Carreras Leukemia Research Institute (IJC), Badalona, 08016 Barcelona, Spain; [email protected] 3 Centro de Investigacion Biomedica en Red Cancer (CIBERONC), 28029 Madrid, Spain 4 Institucio Catalana de Recerca I Estudis Avancats (ICREA), 08016 Barcelona, Spain 5 Physiological Sciences Department, School of Medicine and Health Sciences, University of Barcelona (UB), 08016 Barcelona, Spain 6 Institute of Agri-Food and Life Sciences Agro-Health, Hellenic Mediterranean University, 71003 Crete, Greece * Correspondence: [email protected]; Tel.: +30-694-248-5045 Simple Summary: Our research focuses in the elucidation of the nature of circulating cell-free DNA (ccfDNA) as a biological entity and its exploitation as a liquid biopsy biomaterial. Working on breast cancer, it became clear that although a promising biosource, its clinical exploitation is burdened mainly by gaps in knowledge about its biology and specific characteristics. The current review covers multiple aspects of ccfDNA in breast cancer. We cover key issues such as quantity, integrity, releasing structures, methylation specific changes, release mechanisms, biological role. Machine learning approaches for analyzing ccfDNA-generated data to produce classifiers for clinical use are also discussed. Citation: Panagopoulou, M.; Esteller, M.; Chatzaki, E. Circulating Cell-Free Abstract: Breast cancer (BC) is a leading cause of death between women. Mortality is significantly DNA in Breast Cancer: Searching for raised due to drug resistance and metastasis, while personalized treatment options are obstructed Hidden Information towards by the limitations of conventional biopsy follow-up. -

Transcriptome Analysis Using Next-Generation Sequencing
Available online at www.sciencedirect.com Transcriptome analysis using next-generation sequencing Kai-Oliver Mutz, Alexandra Heilkenbrinker, Maren Lo¨ nne, Johanna-Gabriela Walter and Frank Stahl Up to date research in biology, biotechnology, and medicine more than 15 years and millions of dollars, today it is requires fast genome and transcriptome analysis technologies possible to sequence it in about eight days for approxi- for the investigation of cellular state, physiology, and activity. mately $100 000 [4]. The important technological devel- Here, microarray technology and next generation sequencing opments, summarized under the term next-generation of transcripts (RNA-Seq) are state of the art. Since microarray sequencing, are based on the sequencing-by-synthesis technology is limited towards the amount of RNA, the (SBS) technology called pyrosequencing. The transcrip- quantification of transcript levels and the sequence tomics variant of pyrosequencing technology is called information, RNA-Seq provides nearly unlimited possibilities in short-read massively parallel sequencing or RNA-Seq modern bioanalysis. This chapter presents a detailed [5]. In recent years, RNA-Seq is rapidly emerging as description of next-generation sequencing (NGS), describes the major quantitative transcriptome profiling system the impact of this technology on transcriptome analysis and [6,7]. Different companies developed different sequen- explains its possibilities to explore the modern RNA world. cing platforms all based on variations of pyrosequencing. Address Next-generation sequencing Leibniz Universita¨ t Hannover, Institute for Technical Chemistry, To achieve sequence information, various methods are Callinstrasse 5, 30167 Hannover, Germany well established today. The classical dideoxy method, Corresponding author: Stahl, Frank ([email protected]) invented by Friedrich Sanger in the 1970th uses an enzymatic reaction. -

An Introduction to Next-Generation Sequencing Technology
An introduction to Next-Generation Sequencing Technology www.illumina.com/technology/next-generation-sequencing.html For Research Use Only. Not for use in diagnostic procedures. Table of Contents Table of Contents 2 I. Welcome to Next-Generation Sequencing 3 a. The Evolution of Genomic Science 3 b. The Basics of NGS Chemistry 4 c. Advances in Sequencing Technology 5 Paired-End Sequencing 5 Tunable Coverage and Unlimited Dynamic Range 6 Advances in Library Preparation 6 Multiplexing 7 Flexible, Scalable Instrumentation 7 II. NGS Methods 8 a. Genomics 8 Whole-Genome Sequencing 8 Exome Sequencing 8 De novo Sequencing 9 Targeted Sequencing 9 b. Transcriptomics 11 Total RNA and mRNA Sequencing 11 Targeted RNA Sequencing 11 Small RNA and Noncoding RNASequencing 11 c. Epigenomics 12 Methylation Sequencing 12 ChIP Sequencing 12 Ribosome Profiling 12 III. Illumina DNA-to-Data NGS Solutions 13 a. The Illumina NGS Workflow 13 b. Integrated Data Analysis 13 IV. Glossary 14 V. References 15 For Research Use Only. Not for use in diagnostic procedures. I. Welcome to Next-Generation Sequencing a. The Evolution of Genomic Science DNA sequencing has come a long way since the days of two-dimensional chromatography in the 1970s. With the advent of the Sanger chain termination method1 in 1977, scientists gained the ability to sequence DNA in a reliable, reproducible manner. A decade later, Applied Biosystems introduced the first automated, capillary electrophoresis (CE)-based sequencing instruments,the AB370 in 1987 and the AB3730xl in 1998,instruments that became the primary workhorses for the NIH-led and Celera-led Human Genome Projects.2 While these “first-generation” instruments were considered high throughput for their time, the Genome Analyzer emerged in 2005 and took sequencing runs from 84 kilobase (kb) per run to 1 gigabase (Gb) per run.3 The short read, massively parallel sequencing technique was a fundamentally different approach that revolutionized sequencing capabilities and launched the “next generation” in genomic science. -

Blood Contains Circulating Cell Free Respiratory Competent Mitochondria
Blood contains circulating cell free respiratory competent mitochondria Zahra Al Amir Dache, Amaëlle Otandault, Rita Tanos, Brice Pastor, Romain Meddeb, Cynthia Sanchez, Giuseppe Arena, Laurence Lasorsa, E Bennett, Thierry Grange, et al. To cite this version: Zahra Al Amir Dache, Amaëlle Otandault, Rita Tanos, Brice Pastor, Romain Meddeb, et al.. Blood contains circulating cell free respiratory competent mitochondria: Blood contains extracellular mito- chondria. FASEB Journal, Federation of American Society of Experimental Biology, 2020, 34 (3), pp.3616-3630. 10.1096/fj.201901917RR. hal-02504546 HAL Id: hal-02504546 https://hal.archives-ouvertes.fr/hal-02504546 Submitted on 10 Mar 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Blood contains circulating cell free respiratory competent mitochondria Running Title: Blood contains extracellular mitochondria Zahra Al Amir Dache1, Amaëlle Otandault1, Rita Tanos1, Brice Pastor1, Romain Meddeb1, Cynthia Sanchez1, Giuseppe Arena2, Laurence Lasorsa1, E. Andrew Bennett3, Thierry Grange3, Safia El Messaoudi1, Thibault Mazard1, Corinne Prevostel1, and Alain R. Thierry1* 1 IRCM, Institut de Recherche en Cancérologie de Montpellier, INSERM U1194, Université de Montpellier, Institut régional du Cancer de Montpellier, Montpellier, F-34298, France 2 Department of Developmental and Stem Cell Biology, Institut Pasteur, CNRS, Paris, France.