Molecular Systematics of the Cactaceae
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cop15 Prop. 28
CoP15 Prop. 28 CONVENTION ON INTERNATIONAL TRADE IN ENDANGERED SPECIES OF WILD FAUNA AND FLORA ____________________ Fifteenth meeting of the Conference of the Parties Doha (Qatar), 13-25 March 2010 CONSIDERATION OF PROPOSALS FOR AMENDMENT OF APPENDICES I AND II A. Proposal Delist Euphorbia misera from Appendix II. B. Proponent Mexico and the United States of America* C. Supporting statement 1. Taxonomy 1.1 Class: Magnoliophyta 1.2 Order: Magnoliopsida 1.3 Family: Euphorbiaceae 1.4 Genus, species or subspecies, including author and year: Euphorbia misera Benth. 1.5 Scientific synonyms: Euphorbia benedicta, Trichosterigma benedictum, T. miserum 1.6 Common names: English: cliff spurge, Saint Benedict spurge French: Spanish: hamácj, jumetón, lechosa, golondrina 1.7 Code numbers: 2. Overview Euphorbia misera, native to Mexico and the United States of America, has been listed in CITES Appendix II since 1975. According to CITES trade data, international trade does not appear to be a factor affecting the status of this species. We are proposing to delete this species from the CITES Appendices. Since listing, there has been minimal CITES-recorded international trade (1 shipment of 5 artificially propagated specimens from the United States in the 1990s). The species is intrinsically vulnerable to extinction due to its limited and fragmented distribution and low reproductive output. Euphorbia misera is used medicinally in Mexico, which use appears to be highly localized. The species is known in commercial cultivation in the United States, where there is domestic * The geographical designations employed in this document do not imply the expression of any opinion whatsoever on the part of the CITES Secretariat or the United Nations Environment Programme concerning the legal status of any country, territory, or area, or concerning the delimitation of its frontiers or boundaries. -
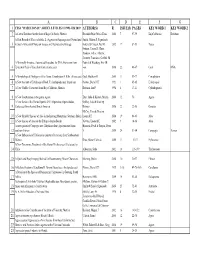
Haseltonia Articles and Authors.Xlsx
ABCDEFG 1 CSSA "HASELTONIA" ARTICLE TITLES #1 1993–#26 2019 AUTHOR(S) R ISSUE(S) PAGES KEY WORD 1 KEY WORD 2 2 A Cactus Database for the State of Baja California, Mexico Resendiz Ruiz, María Elena 2000 7 97-99 BajaCalifornia Database A First Record of Yucca aloifolia L. (Agavaceae/Asparagaceae) Naturalized Smith, Gideon F, Figueiredo, 3 in South Africa with Notes on its uses and Reproductive Biology Estrela & Crouch, Neil R 2012 17 87-93 Yucca Fotinos, Tonya D, Clase, Teodoro, Veloz, Alberto, Jimenez, Francisco, Griffith, M A Minimally Invasive, Automated Procedure for DNA Extraction from Patrick & Wettberg, Eric JB 4 Epidermal Peels of Succulent Cacti (Cactaceae) von 2016 22 46-47 Cacti DNA 5 A Morphological Phylogeny of the Genus Conophytum N.E.Br. (Aizoaceae) Opel, Matthew R 2005 11 53-77 Conophytum 6 A New Account of Echidnopsis Hook. F. (Asclepiadaceae: Stapeliae) Plowes, Darrel CH 1993 1 65-85 Echidnopsis 7 A New Cholla (Cactaceae) from Baja California, Mexico Rebman, Jon P 1998 6 17-21 Cylindropuntia 8 A New Combination in the genus Agave Etter, Julia & Kristen, Martin 2006 12 70 Agave A New Series of the Genus Opuntia Mill. (Opuntieae, Opuntioideae, Oakley, Luis & Kiesling, 9 Cactaceae) from Austral South America Roberto 2016 22 22-30 Opuntia McCoy, Tom & Newton, 10 A New Shrubby Species of Aloe in the Imatong Mountains, Southern Sudan Leonard E 2014 19 64-65 Aloe 11 A New Species of Aloe on the Ethiopia-Sudan Border Newton, Leonard E 2002 9 14-16 Aloe A new species of Ceropegia sect. -

"National List of Vascular Plant Species That Occur in Wetlands: 1996 National Summary."
Intro 1996 National List of Vascular Plant Species That Occur in Wetlands The Fish and Wildlife Service has prepared a National List of Vascular Plant Species That Occur in Wetlands: 1996 National Summary (1996 National List). The 1996 National List is a draft revision of the National List of Plant Species That Occur in Wetlands: 1988 National Summary (Reed 1988) (1988 National List). The 1996 National List is provided to encourage additional public review and comments on the draft regional wetland indicator assignments. The 1996 National List reflects a significant amount of new information that has become available since 1988 on the wetland affinity of vascular plants. This new information has resulted from the extensive use of the 1988 National List in the field by individuals involved in wetland and other resource inventories, wetland identification and delineation, and wetland research. Interim Regional Interagency Review Panel (Regional Panel) changes in indicator status as well as additions and deletions to the 1988 National List were documented in Regional supplements. The National List was originally developed as an appendix to the Classification of Wetlands and Deepwater Habitats of the United States (Cowardin et al.1979) to aid in the consistent application of this classification system for wetlands in the field.. The 1996 National List also was developed to aid in determining the presence of hydrophytic vegetation in the Clean Water Act Section 404 wetland regulatory program and in the implementation of the swampbuster provisions of the Food Security Act. While not required by law or regulation, the Fish and Wildlife Service is making the 1996 National List available for review and comment. -

Caryophyllales 2018 Instituto De Biología, UNAM September 17-23
Caryophyllales 2018 Instituto de Biología, UNAM September 17-23 LOCAL ORGANIZERS Hilda Flores-Olvera, Salvador Arias and Helga Ochoterena, IBUNAM ORGANIZING COMMITTEE Walter G. Berendsohn and Sabine von Mering, BGBM, Berlin, Germany Patricia Hernández-Ledesma, INECOL-Unidad Pátzcuaro, México Gilberto Ocampo, Universidad Autónoma de Aguascalientes, México Ivonne Sánchez del Pino, CICY, Centro de Investigación Científica de Yucatán, Mérida, Yucatán, México SCIENTIFIC COMMITTEE Thomas Borsch, BGBM, Germany Fernando O. Zuloaga, Instituto de Botánica Darwinion, Argentina Victor Sánchez Cordero, IBUNAM, México Cornelia Klak, Bolus Herbarium, Department of Biological Sciences, University of Cape Town, South Africa Hossein Akhani, Department of Plant Sciences, School of Biology, College of Science, University of Tehran, Iran Alexander P. Sukhorukov, Moscow State University, Russia Michael J. Moore, Oberlin College, USA Compilation: Helga Ochoterena / Graphic Design: Julio C. Montero, Diana Martínez GENERAL PROGRAM . 4 MONDAY Monday’s Program . 7 Monday’s Abstracts . 9 TUESDAY Tuesday ‘s Program . 16 Tuesday’s Abstracts . 19 WEDNESDAY Wednesday’s Program . 32 Wednesday’s Abstracs . 35 POSTERS Posters’ Abstracts . 47 WORKSHOPS Workshop 1 . 61 Workshop 2 . 62 PARTICIPANTS . 63 GENERAL INFORMATION . 66 4 Caryophyllales 2018 Caryophyllales General program Monday 17 Tuesday 18 Wednesday 19 Thursday 20 Friday 21 Saturday 22 Sunday 23 Workshop 1 Workshop 2 9:00-10:00 Key note talks Walter G. Michael J. Moore, Berendsohn, Sabine Ya Yang, Diego F. Registration -

Copyright Notice
Copyright Notice This electronic reprint is provided by the author(s) to be consulted by fellow scientists. It is not to be used for any purpose other than private study, scholarship, or research. Further reproduction or distribution of this reprint is restricted by copyright laws. If in doubt about fair use of reprints for research purposes, the user should review the copyright notice contained in the original journal from which this electronic reprint was made. Journal of Vegetation Science 7: 667-680, 1996 © IAVS; Opulus Press Uppsala. Printed in Sweden - Biogeographic patterns of Argentine cacti - 667 Species richness of Argentine cacti: A test of biogeographic hypotheses Mourelle, Cristina & Ezcurra, Exequiel Centro de Ecología, UNAM, Apartado Postal 70-275, 04510 - Mexico, D.F., Mexico; Fax +52 5 6228995; E-mail [email protected] Abstract. Patterns of species richness are described for 50 cacti and (e) pereskioid cacti. Columnar species have columnar, 109 globose and 50 opuntioid cacti species in 318 column-like stems with ribs, formed by an arrangement grid cells (1°×1°) covering Argentina. Biological richness of the areoles in longitudinal rows. These species have hypotheses were tested by regressing 15 environmental parallel vascular bundles, separated by succulent paren- descriptors against species richness in each group. We also chyma, sometimes fusing towards a woody base in the included the collection effort (estimated as the logarithm of the number of herbarium specimens collected in each cell) to adults. We broadly considered as columnar cacti: estimate the possible error induced by underrepresentation in candelabriform arborescent species, unbranched erect certain cells. -

Barbed-Wire Cactus Cereus Tetragonus Also Known As: Acanthocereus Tetragonus Rating: 0.0 ( 0 Votes)
Barbed-wire Cactus Cereus tetragonus Also known as: Acanthocereus tetragonus Rating: 0.0 ( 0 votes) This description is for Barbed-wire Cactus (Cereus tetragonus): Barbed-wire Cactus. Surrounding desert prisons for years to come. Arguably, the most common cactus to have on your office desk, never mind at home, Cereus tetragonus is a dwarf and rather tender perennial, usually potted and grown under glass. Its distinctive three or five-sided stem is mid-green in colour with fine, brown spines growing from small white, hairy tufts along its edges, thus earning it its common name, the Barbed-wire Cactus. Numerous smaller stems appear over time, while the nocturnal yellow or pink blossoms are rarely seen. Though suited to pots and containers, it grows happily in gravel beds and rockeries. Water regularly when growing, but keep dry in winter and apply low-nitrogen fertilizer for extra nourishment. Find Barbed-wire Cactus in our Shop! Free shipping from € 50! Plant Environment Usage Known dangers? Acidity Standard category no Acidic Cacti & succulents Neutral Cacti Alkaline Height [m] Hardiness zone Grown for 0.6 - 0.9 Z10-11 Attractive flowers and foliage Plant Environment Usage Spread [m] Heat zone Creative category 0.3 - 0.4 unknown Kid Approved For Beginners Colours Dominant flower colour Winter temperatures [°C] Garden type White -1 - 4 Indoor or winter garden Containers Flower Fragrance Heat days Gardening expertise No, neutral please 0 beginner Flowering seasons Moisture Time to reach full size Early summer well-drained up to 20 years Mid summer Late summer Early autumn Foliage in spring Soil type Green sandy loams Foliage in summer Sun requirements Green Full sun Foliage in Autumn Exposure Green Sheltered Foliage in winter Green Propagation methods seed root cuttings Growth habit Erect . -

Reproductive Biology, Hybridization, and Flower Visitors of Rare Sclerocactus Taxa in Utah's Uintah Basin
Western North American Naturalist Volume 70 Number 3 Article 10 10-11-2010 Reproductive biology, hybridization, and flower visitors of rare Sclerocactus taxa in Utah's Uintah Basin Vincent J. Tepedino Utah State University, Logan, [email protected] Terry L. Griswold Utah State University, Logan, [email protected] William R. Bowlin Utah State University, Logan Follow this and additional works at: https://scholarsarchive.byu.edu/wnan Recommended Citation Tepedino, Vincent J.; Griswold, Terry L.; and Bowlin, William R. (2010) "Reproductive biology, hybridization, and flower visitors of rare Sclerocactus taxa in Utah's Uintah Basin," Western North American Naturalist: Vol. 70 : No. 3 , Article 10. Available at: https://scholarsarchive.byu.edu/wnan/vol70/iss3/10 This Article is brought to you for free and open access by the Western North American Naturalist Publications at BYU ScholarsArchive. It has been accepted for inclusion in Western North American Naturalist by an authorized editor of BYU ScholarsArchive. For more information, please contact [email protected], [email protected]. Western North American Naturalist 70(3), © 2010, pp. 377–386 REPRODUCTIVE BIOLOGY, HYBRIDIZATION, AND FLOWER VISITORS OF RARE SCLEROCACTUS TAXA IN UTAH’S UINTAH BASIN Vincent J. Tepedino1,2, Terry L. Griswold1, and William R. Bowlin1,3 ABSTRACT.—We studied the mating system and flower visitors of 2 threatened species of Sclerocactus (Cactaceae) in the Uintah Basin of eastern Utah—an area undergoing rapid energy development. We found that both S. wetlandicus and S. brevispinus are predominantly outcrossed and are essentially self-incompatible. A third presumptive taxon (unde- scribed; here called S. wetlandicus-var1) is fully self-compatible but cannot produce seeds unless the flowers are visited by pollinators. -

Acanthocereus Tetragonus SCORE: 16.0 RATING: High Risk (L.) Hummelinck
TAXON: Acanthocereus tetragonus SCORE: 16.0 RATING: High Risk (L.) Hummelinck Taxon: Acanthocereus tetragonus (L.) Hummelinck Family: Cactaceae Common Name(s): barbed-wire cactus Synonym(s): Acanthocereus occidentalis Britton & Rose chaco Acanthocereus pentagonus (L.) Britton & Rose sword-pear Acanthocereus pitajaya sensu Croizat triangle cactus Cactus pentagonus L. Cactus tetragonus L. Assessor: Chuck Chimera Status: Assessor Approved End Date: 1 Nov 2018 WRA Score: 16.0 Designation: H(HPWRA) Rating: High Risk Keywords: Spiny, Agricultural Weed, Environmental Weed, Dense Thickets, Bird-Dispersed Qsn # Question Answer Option Answer 101 Is the species highly domesticated? y=-3, n=0 n 102 Has the species become naturalized where grown? 103 Does the species have weedy races? Species suited to tropical or subtropical climate(s) - If 201 island is primarily wet habitat, then substitute "wet (0-low; 1-intermediate; 2-high) (See Appendix 2) High tropical" for "tropical or subtropical" 202 Quality of climate match data (0-low; 1-intermediate; 2-high) (See Appendix 2) High 203 Broad climate suitability (environmental versatility) y=1, n=0 y Native or naturalized in regions with tropical or 204 y=1, n=0 y subtropical climates Does the species have a history of repeated introductions 205 y=-2, ?=-1, n=0 y outside its natural range? 301 Naturalized beyond native range y = 1*multiplier (see Appendix 2), n= question 205 y 302 Garden/amenity/disturbance weed n=0, y = 1*multiplier (see Appendix 2) n 303 Agricultural/forestry/horticultural weed n=0, y -

Bio 308-Course Guide
COURSE GUIDE BIO 308 BIOGEOGRAPHY Course Team Dr. Kelechi L. Njoku (Course Developer/Writer) Professor A. Adebanjo (Programme Leader)- NOUN Abiodun E. Adams (Course Coordinator)-NOUN NATIONAL OPEN UNIVERSITY OF NIGERIA BIO 308 COURSE GUIDE National Open University of Nigeria Headquarters 14/16 Ahmadu Bello Way Victoria Island Lagos Abuja Office No. 5 Dar es Salaam Street Off Aminu Kano Crescent Wuse II, Abuja e-mail: [email protected] URL: www.nou.edu.ng Published by National Open University of Nigeria Printed 2013 ISBN: 978-058-434-X All Rights Reserved Printed by: ii BIO 308 COURSE GUIDE CONTENTS PAGE Introduction ……………………………………......................... iv What you will Learn from this Course …………………............ iv Course Aims ……………………………………………............ iv Course Objectives …………………………………………....... iv Working through this Course …………………………….......... v Course Materials ………………………………………….......... v Study Units ………………………………………………......... v Textbooks and References ………………………………........... vi Assessment ……………………………………………….......... vi End of Course Examination and Grading..................................... vi Course Marking Scheme................................................................ vii Presentation Schedule.................................................................... vii Tutor-Marked Assignment ……………………………….......... vii Tutors and Tutorials....................................................................... viii iii BIO 308 COURSE GUIDE INTRODUCTION BIO 308: Biogeography is a one-semester, 2 credit- hour course in Biology. It is a 300 level, second semester undergraduate course offered to students admitted in the School of Science and Technology, School of Education who are offering Biology or related programmes. The course guide tells you briefly what the course is all about, what course materials you will be using and how you can work your way through these materials. It gives you some guidance on your Tutor- Marked Assignments. There are Self-Assessment Exercises within the body of a unit and/or at the end of each unit. -

University of Florida Thesis Or Dissertation Formatting
SYSTEMATICS OF TRIBE TRICHOCEREEAE AND POPULATION GENETICS OF Haageocereus (CACTACEAE) By MÓNICA ARAKAKI MAKISHI A DISSERTATION PRESENTED TO THE GRADUATE SCHOOL OF THE UNIVERSITY OF FLORIDA IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY UNIVERSITY OF FLORIDA 2008 1 © 2008 Mónica Arakaki Makishi 2 To my parents, Bunzo and Cristina, and to my sisters and brother. 3 ACKNOWLEDGMENTS I want to express my deepest appreciation to my advisors, Douglas Soltis and Pamela Soltis, for their consistent support, encouragement and generosity of time. I would also like to thank Norris Williams and Michael Miyamoto, members of my committee, for their guidance, good disposition and positive feedback. Special thanks go to Carlos Ostolaza and Fátima Cáceres, for sharing their knowledge on Peruvian Cactaceae, and for providing essential plant material, confirmation of identifications, and their detailed observations of cacti in the field. I am indebted to the many individuals that have directly or indirectly supported me during the fieldwork: Carlos Ostolaza, Fátima Cáceres, Asunción Cano, Blanca León, José Roque, María La Torre, Richard Aguilar, Nestor Cieza, Olivier Klopfenstein, Martha Vargas, Natalia Calderón, Freddy Peláez, Yammil Ramírez, Eric Rodríguez, Percy Sandoval, and Kenneth Young (Peru); Stephan Beck, Noemí Quispe, Lorena Rey, Rosa Meneses, Alejandro Apaza, Esther Valenzuela, Mónica Zeballos, Freddy Centeno, Alfredo Fuentes, and Ramiro Lopez (Bolivia); María E. Ramírez, Mélica Muñoz, and Raquel Pinto (Chile). I thank the curators and staff of the herbaria B, F, FLAS, LPB, MO, USM, U, TEX, UNSA and ZSS, who kindly loaned specimens or made information available through electronic means. Thanks to Carlos Ostolaza for providing seeds of Haageocereus tenuis, to Graham Charles for seeds of Blossfeldia sucrensis and Acanthocalycium spiniflorum, to Donald Henne for specimens of Haageocereus lanugispinus; and to Bernard Hauser and Kent Vliet for aid with microscopy. -

Species Classification and Nomenclature by Norbert Leist and Andrea Jonitz Prof
ISTA Purity Seminar 15. June 2009 Zürich TlTools for seed identifi cati on species classification and nomenclature by Norbert Leist and Andrea Jonitz Prof. Dr. Norbert Leist Dr. Andrea Jonitz Brahmsstr.25 LTZ Augustenberg 76669 Bad Schönborn Neßlerstr.23 Germany 76227 Karlsruhe [email protected] Germany [email protected] Aquilegia vulgaris, Variation Variation • Variation is everywhere in biological systems. Natural variation at the population level is usualy not continuous, but occurs in discrete units or taxa. Easily the most important taxonomic level is the species because it is often the smallest clearly recognizable and discrete set of populations. • Understanding how species form and how to recognize them have been major challenges to systematists. The variation in one population becomes interrupted, the way to a split into two species strong hairy nearly glabrous Variation on species • Sources of variation: MttiMutation Recombination Independent assortment of the chromosomes Random genetic drift Selection Conservation of species characteristics avoiding gene flow Isolating barriers: temporal (seasonal, diurnal) habitat (wet, dry; calceous, silicious) floral (structural, behavioral eg. adaptations for pollinators) reproductive mode (self fertilisation, agamospery) incompatibility (pollen, seeds) hybrid inviability hybrid floral isolation hybrid sterility hybrid break down Iris germanica Iris sibirica Isolation by habitat Definition of „species“ is not easy A species is the smallest aggregation of populations -

Evolutionary History of Floral Key Innovations in Angiosperms Elisabeth Reyes
Evolutionary history of floral key innovations in angiosperms Elisabeth Reyes To cite this version: Elisabeth Reyes. Evolutionary history of floral key innovations in angiosperms. Botanics. Université Paris Saclay (COmUE), 2016. English. NNT : 2016SACLS489. tel-01443353 HAL Id: tel-01443353 https://tel.archives-ouvertes.fr/tel-01443353 Submitted on 23 Jan 2017 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. NNT : 2016SACLS489 THESE DE DOCTORAT DE L’UNIVERSITE PARIS-SACLAY, préparée à l’Université Paris-Sud ÉCOLE DOCTORALE N° 567 Sciences du Végétal : du Gène à l’Ecosystème Spécialité de Doctorat : Biologie Par Mme Elisabeth Reyes Evolutionary history of floral key innovations in angiosperms Thèse présentée et soutenue à Orsay, le 13 décembre 2016 : Composition du Jury : M. Ronse de Craene, Louis Directeur de recherche aux Jardins Rapporteur Botaniques Royaux d’Édimbourg M. Forest, Félix Directeur de recherche aux Jardins Rapporteur Botaniques Royaux de Kew Mme. Damerval, Catherine Directrice de recherche au Moulon Président du jury M. Lowry, Porter Curateur en chef aux Jardins Examinateur Botaniques du Missouri M. Haevermans, Thomas Maître de conférences au MNHN Examinateur Mme. Nadot, Sophie Professeur à l’Université Paris-Sud Directeur de thèse M.