Molecular Taxonomy of the Alternaria and Ulocladium Species from Humans and Their Identification in the Routine Laboratory
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Symptomology, Biology and Management of Alternaria Leaf Spot
The Pharma Innovation Journal 2021; 10(6): 264-268 ISSN (E): 2277- 7695 ISSN (P): 2349-8242 NAAS Rating: 5.23 Symptomology, biology and management of Alternaria TPI 2021; 10(6): 264-268 © 2021 TPI leaf spot of mustard (Brassica spp.) www.thepharmajournal.com Received: 24-04-2021 Accepted: 30-05-2021 Ramesh Kumar and Poonam Shete Ramesh Kumar Department of Plant Pathology, Abstract School of Agriculture, Lovely Oilseed Brassica spp. is one of the most important diseases of oilseed crop in the world. Rapeseed Professional University, mustard are susceptible to a number of diseases which is caused by the living (biotic) pathogen. It is also Phagwara, Punjab, India known as Alternaria black spot diseases surrounded with yellow colours on the leaves which is known to be the most destructive diseases in the world. This disease is generally characterised by the different Poonam Shete names which are as follows, Alternaria brassica, Alternaria brassicola and Alternaria raphani. Department of Plant Pathology, Alternaria leaf spot pathogen produces lesion around the leaves, stem, and the Silique which cause School of Agriculture, Lovely reduction in defoliation. These pathogens are seed borne, soil borne, and airborne diseases. Alternaria Professional University, leaf spot diseases caused by the heavy rainfall and the weather with the highest diseases incidence. The Phagwara, Punjab, India Conidia, age of the host plant is also responsible for severity of the diseases. This disease is more 0 prominent during the summer seasons where the temperature falls 27- 28 C. This paper also determines the development of Alternaria leaf blightin Mustard crop in relation to the pathogen such as taxonomy, biology, epidemiology and their management through biological, chemical, cultural and botanical approaches. -

Management of Alternaria Leaf and Fruit Spot in Apples
Management of Alternaria leaf and fruit spot in apples Christine Horlock QLD Department of Primary Industries and Fisheries Project Number: AP02011 AP02011 This report is published by Horticulture Australia Ltd to pass on information concerning horticultural research and development undertaken for the apple and pear industry. The research contained in this report was funded by Horticulture Australia Ltd with the financial support of the apple and pear industry. All expressions of opinion are not to be regarded as expressing the opinion of Horticulture Australia Ltd or any authority of the Australian Government. The Company and the Australian Government accept no responsibility for any of the opinions or the accuracy of the information contained in this report and readers should rely upon their own enquiries in making decisions concerning their own interests. ISBN 0 7341 1256 4 Published and distributed by: Horticulture Australia Ltd Level 1, 50 Carrington Street Sydney NSW 2000 T: (02) 8295 2300 F: (02) 8295 2399 E: [email protected] © Copyright 2006 FINAL REPORT AP02011 (28 February 2006) Management of Alternaria leaf and fruit spot in apples Christine M. Horlock et al. Department of Primary Industries and Fisheries, Queensland AP02011 Ms Christine M. Horlock Plant Pathologist, Delivery Department of Primary Industries and Fisheries, Queensland Applethorpe Research Station PO Box 501 STANTHORPE QLD 4380 Dr Shane Hetherington Deciduous Fruits Pathologist NSW Department of Primary Industries, Agriculture Orange Agricultural Institute Forest Road ORANGE NSW 2800 Mr Shane Dullahide Former Senior Experimentalist, Delivery Department of Primary Industries and Fisheries, Queensland Applethorpe Research Station PO Box 501 STANTHORPE QLD 4380 This document is the final report for the APAL / HAL funded project “Management of Alternaria leaf and fruit spot in apples”, and as such contains the details of all scientific work carried out in this project. -

Nature and Effect of Alternaria Spp. Complex from Wheat Grain on Germination and Disease Transmission
Pak. J. Bot., 45(5): 1817-1824, 2013. NATURE AND EFFECT OF ALTERNARIA SPP. COMPLEX FROM WHEAT GRAIN ON GERMINATION AND DISEASE TRANSMISSION ANALÍA E. PERELLÓ1,2* AND SILVINA LARRÁN1 1CIDEFI (Centro de Investigaciones de Fitopatología) y Cátedra de Fitopatología 2CONICET-Facultad de Ciencias Agrarias y Forestales de la Universidad Nacional de La Plata, Calle 60 y 119 (1900) La Plata, Buenos Aires, Argentina. *Corresponding author’s e-mail: anaperello2@ yahoo.com.ar Abstract Diseases caused by Alternaria sp. are among the most common diseases of crops throughout the world. Alternaria sp. is a common component of the flora of wheat seed. Although isolation of Alternaria sp. from wheat (Triticum aestivum) seed has been reported in Argentina, development of the Alternaria blight in plants from infected seeds has not been demonstrated experimentally. Seed transmission of strains belonging to Alternaria tenuissima, A. alternata, A. infectoria, A. triticina, A. chlamydospora and related genera like Embellisia and Ulocladium sp. on wheat were investigated in the Argentinean growing area, on wheat cultivars Klein Escorpión and Buck Poncho. A. tenuissima was the dominant fungus in black pointed kernels. Transmission of all 42 seed-borne members of Alternaria complex from seeds to seedlings artificially inoculated was detected by trays seedling symptoms test. Among the fungi tested most isolates of Alternaria, Embellisia sp. and Ulocladium sp. produced distinct seed rot and seedling infection symptoms. This confirmed the seed-borne nature of these fungi. In each wheat cultivar tested inoculated seeds appreciably reduced their germination. The emerging coleoptile is externally infected by hyphal growth from the infected pericarp. -

In Vitro Evaluation of Plant Extracts and Bio- Agents Against Alternaria
International Journal of Chemical Studies 2018; 6(2): 504-507 P-ISSN: 2349–8528 E-ISSN: 2321–4902 IJCS 2018; 6(2): 504-507 In vitro evaluation of plant extracts and bio- © 2018 IJCS Received: 04-01-2018 agents against Alternaria tenuissima (Fr.) keissl Accepted: 05-02-2018 causing leaf blight of kodo millet K Hariprasad Department of Plant Pathology, UAS, GKVK, Bengaluru - 65 K Hariprasad, A Nagaraja, Suresh Patil and Rakesha *Project Coordinating Unit (Small Millets), ICAR, GKVK, Abstract Bengaluru, Karnataka, India Kodo millet (Paspalum scrobiculatum L.) is nutritionally important millet. Leaf blight has been a major production constraint and fungicidal sprays for the management of any disease on this crop may not be A Nagaraja economically viable and feasible as the farmers cultivating the crop are resource poor and the crop is less Department of Plant Pathology, remunerative, but is important in tribal and rainfed agriculture. Hence, botanicals and bio-agents were UAS, GKVK, Bengaluru - 65 *Project Coordinating Unit evaluated in vitro against Alternaria tenuissima the cause of leaf blight. Ten plant extracts and 14 bio- (Small Millets), ICAR, GKVK, agents were evaluated following dual culture. The results revealed 100 per cent inhibition of the mycelial Bengaluru, Karnataka, India growth of A. tenuissima by Eucalyptus sp. and Clerodendron infortunatum at 7.5 and 10.0 per cent concentrations. Among the fungal bio control agents tested, maximum inhibition (100 %) was recorded Suresh Patil in Trichoderma harzianum (NBAIR), followed by T. viride (81.38 %); whereas the bacterial bio agent Department of Plant Pathology, Bacillus amyloliquefaciens (P-42) showed only 77.40 % inhibition of mycelial growth revealing that UAS, GKVK, Bengaluru - 65 fungal antagonists were more effective than the bacterial antagonists. -

1 Etiology, Epidemiology and Management of Fruit Rot Of
Etiology, Epidemiology and Management of Fruit Rot of Deciduous Holly in U.S. Nursery Production Dissertation Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Shan Lin Graduate Program in Plant Pathology The Ohio State University 2018 Dissertation Committee Dr. Francesca Peduto Hand, Advisor Dr. Anne E. Dorrance Dr. Laurence V. Madden Dr. Sally A. Miller 1 Copyrighted by Shan Lin 2018 2 Abstract Cut branches of deciduous holly (Ilex spp.) carrying shiny and colorful fruit are popularly used for holiday decorations in the United States. Since 2012, an emerging disease causing the fruit to rot was observed across Midwestern and Eastern U.S. nurseries. A variety of other symptoms were associated with the disease, including undersized, shriveled, and dull fruit, as well as leaf spots and early plant defoliation. The disease causal agents were identified by laboratory processing of symptomatic fruit collected from nine locations across four states over five years by means of morphological characterization, multi-locus phylogenetic analyses and pathogenicity assays. Alternaria alternata and a newly described species, Diaporthe ilicicola sp. nov., were identified as the primary pathogens associated with the disease, and A. arborescens, Colletotrichum fioriniae, C. nymphaeae, Epicoccum nigrum and species in the D. eres species complex were identified as minor pathogens in this disease complex. To determine the sources of pathogen inoculum in holly fields, and the growth stages of host susceptibility to fungal infections, we monitored the presence of these pathogens in different plant tissues (i.e., dormant twigs, mummified fruit, leaves and fruit), and we studied inoculum dynamics and assessed disease progression throughout the growing season in three Ohio nurseries exposed to natural inoculum over two consecutive years. -

An Unusual Haemoid Fungi: Ulocladium, As a Cause Of
Volume 2 Number 2 (June 2010) 95-97 CASE Report An unusual phaeoid fungi: Ulocladium, as a cause of chronic allergic fungal sinusitis Kaur R, Wadhwa A1,Gulati A2, Agrawal AK2 1Department of Microbiology. 2Department of Otorhinolaryngology, Maulana Azad Medical College, New Delhi, India. Received: April 2010, Accepted: May 2010. ABSTRACT Allergic fungal sinusitis (AFS) has been recognized as an important cause of chronic sinusitis commonly caused by Aspergillus spp. and various dematiaceous fungi like Bipolaris, Alternaria, Curvalaria, and etc. Ulocladium botrytis is a non pathogenic environmental dematiaceous fungi, which has been recently described as a human pathogen. Ulocladium has never been associated with allergic fungal sinusitis but it was identified as an etiological agent of AFS in a 35 year old immunocompetent female patient presenting with chronic nasal obstruction of several months duration to our hospital. The patient underwent FESS and the excised polyps revealed Ulocladium as the causative fungal agent. Keywords: Ulocladium, Phaeoid Fungi, Chronic Sinusitis. INTRODUCTION CASE REPORT Chronic allergic sinusitis is a common condition A 35 year old female patient presented to the ENT responsible for the development of nasal polyps, out patient department of the Lok Nayak Hospital, described as abnormal lesions that emanate from Delhi, with chronic nasal obstruction, excessive any portion of the nasal mucosa or Para nasal sneezing, nasal discharge and frontal headache since sinuses. They are commonly located in the middle several months. Nasal obstruction was of gradual meatus and ethmoid sinus and are present in onset, non progressive, more on the left side than 1-4% of the population (1). -

Genetic Variability in the Pistachio Late Blight Fungus, Alternaria Alternata
Mycol. Res. 105 (3): 300–306 (March 2001). Printed in the United Kingdom. 300 Genetic variability in the pistachio late blight fungus, Alternaria alternata Mallikarjuna K. ARADHYA*, Helen M. CHAN and Dan E. PARFITT Department of Pomology, University of California, One Shields Avenue, Davis, CA 95616, USA. E-mail: aradhya!ucdavis.edu Received 15 December 1999; accepted 21 August 2000 Genetic variation in the pistachio late blight fungus, Alternaria alternata, was investigated by restriction fragment length polymorphism (RFLP) in the rDNA region. Southern hybridization of EcoRI, HindIII, and XbaI digested fungal DNA with a RNA probe derived from Alt1, an rDNA clone isolated from a genomic library of the Japanese pear pathotype of A. alternata, revealed 34 different rDNA haplotypes among 56 isolates collected from four central valley locations in California. Analysis of molecular variation revealed a significant amount of genetic diversity within populations (85n8%), with only marginal variation accounting for differentiation among populations (14n2%, ΦST l 0n142). All isolates examined were highly pathogenic. The identity of the four geographic populations sampled was not evident in both cluster and principal component analyses, probably indicating either the selectively neutral nature of rDNA variation or prevalence of widespread gene flow among populations combined with uniform host-selection. INTRODUCTION 1998). Biochemical and molecular markers have been used to demonstrate the existence of multiple strains and to assess The genus Alternaria contains diverse and ubiquitous species of infraspecific variation in A. alternata at the protein and DNA fungi, including aggressive and opportunistic plant pathogens level (Petrunak & Christ 1992, Adachi et al. 1993, Weir et al. -

Alternaria Diseases of Crucifers: Biology, Ecology and Disease Management Alternaria Diseases of Crucifers: Biology, Ecology and Disease Management
Govind Singh Saharan Naresh Mehta Prabhu Dayal Meena Alternaria Diseases of Crucifers: Biology, Ecology and Disease Management Alternaria Diseases of Crucifers: Biology, Ecology and Disease Management Govind Singh Saharan • Naresh Mehta Prabhu Dayal Meena Alternaria Diseases of Crucifers: Biology, Ecology and Disease Management Govind Singh Saharan Naresh Mehta Plant Pathology Plant Pathology CCS Haryana Agricultural University CCS Haryana Agricultural University Hisar , Haryana , India Hisar , Haryana , India Prabhu Dayal Meena Crop Protection Unit ICAR Bharatpur , Rajasthan , India ISBN 978-981-10-0019-5 ISBN 978-981-10-0021-8 (eBook) DOI 10.1007/978-981-10-0021-8 Library of Congress Control Number: 2015958091 Springer Singapore Heidelberg New York Dordrecht London © Springer Science+Business Media Singapore 2016 This work is subject to copyright. All rights are reserved by the Publisher, whether the whole or part of the material is concerned, specifi cally the rights of translation, reprinting, reuse of illustrations, recitation, broadcasting, reproduction on microfi lms or in any other physical way, and transmission or information storage and retrieval, electronic adaptation, computer software, or by similar or dissimilar methodology now known or hereafter developed. The use of general descriptive names, registered names, trademarks, service marks, etc. in this publication does not imply, even in the absence of a specifi c statement, that such names are exempt from the relevant protective laws and regulations and therefore free for general use. The publisher, the authors and the editors are safe to assume that the advice and information in this book are believed to be true and accurate at the date of publication. Neither the publisher nor the authors or the editors give a warranty, express or implied, with respect to the material contained herein or for any errors or omissions that may have been made. -
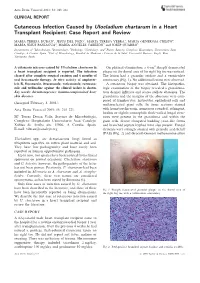
Cutaneous Infection Caused by Ulocladium Chartarum in a Heart Transplant Recipient: Case Report and Review
Acta Derm Venereol 2003; 83: 218–221 CLINICAL REPORT Cutaneous Infection Caused by Ulocladium chartarum in a Heart Transplant Recipient: Case Report and Review MARI´A TERESA DURA´ N1, JESU´ S DEL POZO2, MARI´A TERESA YEBRA3, MARI´A GENEROSA CRESPO4, MARI´A JESU´ S PANIAGUA4, MARI´A ANGELES CABEZO´ N5 and JOSEP GUARRO6 Departments of 1Microbiology, 2Dermatology, 3Pathology, 4Cardiology, and 5Plastic Surgery, Complexo Hospitalario Universitario Juan Canalejo, A Corun˜a, Spain, 6Unit of Microbiology, Facultat de Medicina i Cie`ncies de la Salut, Universitat Rovira i Virgili, Reus, Tarragona, Spain A cutaneous mycoses caused by Ulocladium chartarum in On physical examination, a 6-cm2 sharply demarcated a heart transplant recipient is reported. The infection plaque on the dorsal area of his right big toe was noticed. cleared after complete surgical excision and 6 months of The lesion had a granular surface and a vermiculate oral itraconazole therapy. In vitro activity of amphoter- consistency (Fig. 1). No additional lesions were observed. icin B, fluconazole, itraconazole, voriconazole, ravucona- A cutaneous biopsy was obtained. The histopatho- zole and terbinafine against the clinical isolate is shown. logic examination of the biopsy revealed a granuloma- Key words: dermatomycoses; immunocompromised host; tous dermal infiltrate and scarce stellate abscesses. The skin diseases. granuloma and the margins of the abscesses were com- posed of lymphocytes, histiocytes, epithelioid cells and (Accepted February 3, 2003.) multinucleated giant cells. In tissue sections stained Acta Derm Venereol 2003; 83: 218–221. with hematoxylin-eosin, numerous rounded, refringent, hyaline or slightly eosinophilic thick-walled fungal struc- Ma Teresa Dura´n Valle, Servicio de Microbiologı´a, tures were present in the granuloma and within the Complexo Hospitalario Universitario Juan Canalejo, giant cells. -

Characterization of Alternaria Alternata Isolates Causing Brown Spot of Potatoes in South Africa
Characterization of Alternaria alternata isolates causing brown spot of potatoes in South Africa By Joel Prince Dube Submitted in partial fulfilment of the requirements for the degree of Master in Science (Agriculture) Plant Pathology In the faculty of Natural and Agricultural Sciences Department of Microbiology and Plant Pathology University of Pretoria Pretoria February 2014 © University of Pretoria DECLARATION I, Joel Prince Dube, declare that the thesis, which I hereby submit for the degree Master of Science (Agriculture) Plant Pathology at the University of Pretoria, is my own work and has not been previously submitted by me for a degree at this or any other tertiary institution. Signed: ___________________________ Date: ____________________________ i © University of Pretoria Acknowledgements I would like to extend my heartfelt thanks the contributions of the following: 1. First and foremost, the Almighty God by whose grace I am where I am today. I owe everything to him. 2. My supervisors, Prof. Jacquie van der Waals and Dr. Mariette Truter, for their unwavering support and guidance throughout my Masters journey. 3. Pathology programme @ UP for the opportunity and funding for my studies. 4. Syngenta for funding one of my chapters. 5. Charles Wairuri, Nelisiwe Khumalo, Alain Misse for their help with all my molecular work. 6. Colleagues in greenhouse for all their help, suggestions and contributions throughout my studies. 7. My family and friends for their financial, spiritual and moral support, it is greatly appreciated. ii © University of Pretoria Characterization of Alternaria alternata isolates causing brown spot of potatoes in South Africa By Joel Prince Dube Supervisor : Prof. J. -

Identification and Characterization of Alternaria Species Causing Leaf Spot
Eur J Plant Pathol (2017) 149:401–413 DOI 10.1007/s10658-017-1190-0 Identification and characterization of Alternaria species causing leaf spot on cabbage, cauliflower, wild and cultivated rocket by using molecular and morphological features and mycotoxin production Ilenia Siciliano & Giovanna Gilardi & Giuseppe Ortu & Ulrich Gisi & Maria Lodovica Gullino & Angelo Garibaldi Accepted: 22 February 2017 /Published online: 11 March 2017 # Koninklijke Nederlandse Planteziektenkundige Vereniging 2017 Abstract Alternaria species are common pathogens of Keywords Crucifers . Toxins . Leaf spot . Tenuazonic fruit and vegetables able to produce secondary metabo- acid lites potentially affecting human health. Twenty-nine isolates obtained from cabbage, cauliflower, wild and cultivated rocket were characterized and identified Introduction based on sporulation pattern and virulence; the phylo- β genetic analysis was based on the -tubulin gene. Most Alternaria species are saprophytes and ubiquitous Isolates were identified as A. alternata, A. tenuissima, in the environment, however some are plant pathogenic, A. arborescens, A. brassicicola and A. japonica. inducing diseases on a large variety of economically Pathogenicity was evaluated on plants under greenhouse important crops like cereals, oil-crops, vegetables and conditions. Two isolates showed low level of virulence fruits (Pitt and Hocking, 1997). Most Alternaria spp. on cultivated rocket while the other isolates showed produce chains of conidia with transverse and longitu- medium or high level of virulence. Isolates were also dinal septa with a tapering apical cell. Conidial size, characterized for their mycotoxin production on a mod- presence and size of a beak, the pattern of catenation ified Czapek-Dox medium. Production of the five and longitudinal and transverse septation are key taxo- Alternaria toxins, tenuazonic acid, alternariol, nomic features for this genus (Joly 1964; Ellis 1971 and alternariol monomethyl ether, altenuene and tentoxin 1976, Simmons 1992). -

Studies in Mycology 75: 171–212
STUDIES IN MYCOLOGY 75: 171–212. Alternaria redefined J.H.C. Woudenberg1,2*, J.Z. Groenewald1, M. Binder1 and P.W. Crous1,2,3 1CBS-KNAW Fungal Biodiversity Centre, Uppsalalaan 8, 3584 CT Utrecht, The Netherlands; 2Wageningen University and Research Centre (WUR), Laboratory of Phytopathology, Droevendaalsesteeg 1, 6708 PB Wageningen, The Netherlands; 3Utrecht University, Department of Biology, Microbiology, Padualaan 8, 3584 CH Utrecht, The Netherlands *Correspondence: Joyce H.C. Woudenberg, [email protected] Abstract: Alternaria is a ubiquitous fungal genus that includes saprobic, endophytic and pathogenic species associated with a wide variety of substrates. In recent years, DNA- based studies revealed multiple non-monophyletic genera within the Alternaria complex, and Alternaria species clades that do not always correlate to species-groups based on morphological characteristics. The Alternaria complex currently comprises nine genera and eight Alternaria sections. The aim of this study was to delineate phylogenetic lineages within Alternaria and allied genera based on nucleotide sequence data of parts of the 18S nrDNA, 28S nrDNA, ITS, GAPDH, RPB2 and TEF1-alpha gene regions. Our data reveal a Pleospora/Stemphylium clade sister to Embellisia annulata, and a well-supported Alternaria clade. The Alternaria clade contains 24 internal clades and six monotypic lineages, the assemblage of which we recognise as Alternaria. This puts the genera Allewia, Brachycladium, Chalastospora, Chmelia, Crivellia, Embellisia, Lewia, Nimbya, Sinomyces, Teretispora, Ulocladium, Undifilum and Ybotromyces in synonymy with Alternaria. In this study, we treat the 24 internal clades in the Alternaria complex as sections, which is a continuation of a recent proposal for the taxonomic treatment of lineages in Alternaria.